
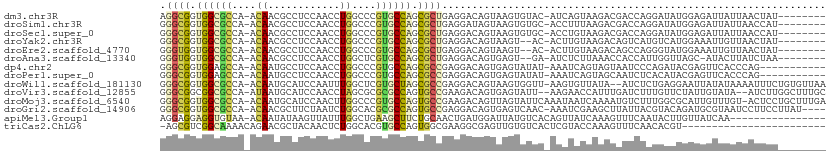
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,717,352 – 10,717,493 |
| Length | 141 |
| Max. P | 0.896152 |

| Location | 10,717,352 – 10,717,459 |
|---|---|
| Length | 107 |
| Sequences | 15 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.18 |
| Shannon entropy | 0.58983 |
| G+C content | 0.59542 |
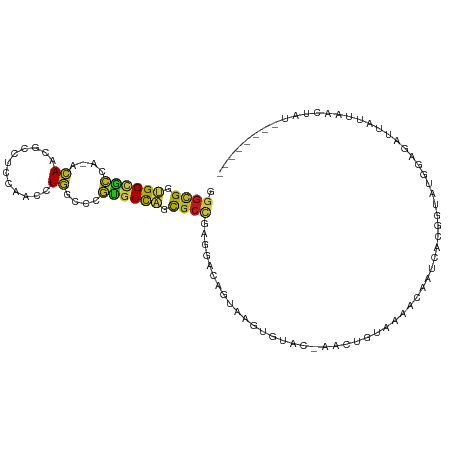
| Mean single sequence MFE | -40.41 |
| Consensus MFE | -27.17 |
| Energy contribution | -27.29 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.28 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10717352 107 + 27905053 UGGUGGCCAACACCUUGGUGGGUGCUUGGGAUCGUUCCCAAGGCGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGACAGUAAGUGUACAUCA----------- (((((((((.(((....)))(((.(((((((....)))))))..((.((((...-....)))).)).))))))))...))))(((((..........)))))......----------- ( -43.30, z-score = -0.23, R) >droSim1.chr3R 16773642 107 - 27517382 UGGUGGCCAACACCUUGGUGGGUGCUUGGGAUCGUUCUCAGGGCGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGAUAGUAAGUGUGCACCU----------- .((((.....))))..((((.(..((((..(((...(((((.((((.((((...-....)))).))....((((....)))))).))))))))..))))..).)))).----------- ( -47.60, z-score = -1.00, R) >droSec1.super_0 9902786 107 - 21120651 UGGUGGCCAACACCUUGGUGGGUGCUUGGGAUCGUUCCCAGGGCGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGACAGUAAGUGUGCACCU----------- ..(.(((((.(((....)))(((.(((((((....)))))))..((.((((...-....)))).)).)))))))))((((((..((((....))))...)).))))..----------- ( -44.70, z-score = 0.36, R) >droYak2.chr3R 13121753 105 - 28832112 UGGUGGCCAACACCUUGGUGGGUGCCUGGGAUCGUUCCCAGGGCGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGACAGUAAGUACACUU------------- ..(.(((((.(((....)))(((.(((((((....)))))))..((.((((...-....)))).)).)))))))))((((....((((....))))..))))....------------- ( -44.70, z-score = -0.33, R) >droEre2.scaffold_4770 10253543 105 + 17746568 UGGUGGCCAACACCUUGGUGGGUGCCUGGGAUCGUUCCCAGGGUGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGACAGUAAGUACACUU------------- ..(.(((((.(((....)))(((.(((((((....))))))).(((.((((...-....)))).))))))))))))((((....((((....))))..))))....------------- ( -45.30, z-score = -0.54, R) >droAna3.scaffold_13340 13723434 105 - 23697760 UGGUGGCCAACACUCUGGUUGGUGCUUGGGAUCGUUCCCAGGGUGGUGGCGCCA-ACAACGCCUCCAACCUGGCUCGUGCCAGCGCUGAGGACAGUGAGUGAAUCU------------- ..........(((((..(((((..(.(((((....)))))((((((.((((...-....)))).)).)))).....)..)))))((((....))))))))).....------------- ( -43.10, z-score = -0.67, R) >dp4.chr2 7799619 109 + 30794189 UGGUGGCCAACACCUUGGUGGGUGCCUGGGAUCGCUCUCAGGGCGGUGGAGCCA-ACAAUGCCUCCAACCUGGCCCGUGCCAGCGCCGAGGACAGUGAGUAUAUAAAUCA--------- .(((.((((...((((((((((..(.(((((....)))))(((((((((((.((-....)).)))).)))..)))))..))..))))))))....)).))......))).--------- ( -40.00, z-score = 0.54, R) >droPer1.super_0 1623045 109 + 11822988 UGGUGGCCAACACCUUGGUGGGUGCCUGGGAUCGCUCUCAGGGCGGUGGAGCCA-ACAAUGCCUCCAACCUGGCCCGUGCCAGCGCCGAGGACAGUGAGUAUAUAAAUCA--------- .(((.((((...((((((((((..(.(((((....)))))(((((((((((.((-....)).)))).)))..)))))..))..))))))))....)).))......))).--------- ( -40.00, z-score = 0.54, R) >droWil1.scaffold_181130 12178541 118 - 16660200 UGGUGGCCAACACUUUGGUUGGUGCUUGGGAUCGCUCACAGGGCGGUGGCGCCA-ACAAUGCAUCCAAUUUGGCUCGUGCUAGCGCCGAGGACAGUAAGUGGUUAAGUGUUAUAAUCUC .(((....(((((((..(((((((((....((((((.....)))))))))))))-)).......(((..(((.((((.((....))))))..)))....)))..)))))))...))).. ( -39.60, z-score = -0.40, R) >droVir3.scaffold_12855 5383803 109 - 10161210 UGGUGGCCAACACUUUAGUUGGUGCUUGGGACCGAUCCCAGGGCGGCGGCGCCA-AUAAUGCAUCCAACCUAGCGCGCGCCAGUGCCGAAGACAGUGAGUA-UUAAGAACC-------- .(((.((((((......)))))).(((((((....)))))))..((((((((..-.................)))).))))...)))..............-.........-------- ( -34.01, z-score = 0.55, R) >droMoj3.scaffold_6540 25002908 111 - 34148556 UGGUGGCCAACACCUUGGUAGGUGCCUGGGAUCGUUCCCAGGGCGGUGGCGCCA-ACAAUGCAUCCAACUUGGCCCGUGCCAGUGCCGAAGACAGUUAGUAUUCAAAUAAUC------- .((((.....))))..(((((((((((((((....))))))((((....)))).-.....)))))....(((((....)))))))))(((.((.....)).)))........------- ( -39.00, z-score = -0.51, R) >droGri2.scaffold_14906 6891121 108 - 14172833 UGGUGGCCAAUACUCUGGUUGGUGCCUGGGAUCGUUCUCAGGGCGGUGGCGCCA-ACAACGCUUCUAAUCUGGCACGCGCCAGUGCCGAGGACAGUGAGUCAACAAAUC---------- ((.((((((...(((..(((((((((....((((((.....)))))))))))))-))..............(((((......)))))))).....)).)))).))....---------- ( -39.20, z-score = -0.61, R) >anoGam1.chrX 17670515 90 + 22145176 UGGUGGCGAAUACGCUGAUCGGCGCGUGGGACCGGUCGCAGGGCGGUGGCGCCA-ACAACGCGUCGAAGCUGGC----GAUGGCGUCGGCGACCG------------------------ .(((((((....))))..((((((((((.((....)).)..((((....)))).-...))))))))).((((((----(....))))))).))).------------------------ ( -41.50, z-score = -0.38, R) >apiMel3.Group1 13435752 100 + 25854376 UGGUUGCAAAUACCCUUAUGGGAGCAUGGGAUCGUAGUCAAGGAGGAGGUGUAA-ACAAUAUAAGUUAUUUGGCUGAAGCUUCUGCAACUGAUGGAUUAUG------------------ .((((((.....(((....))).....((((...((((((((...((..((((.-....))))..)).))))))))....))))))))))...........------------------ ( -22.80, z-score = -0.15, R) >triCas2.ChLG6 6402020 105 - 13544221 UGGUGGCUUCCACCUUGUUGGGUGCAUGGGAUAGGUCCCAAG-CGUCGGCAAAACAGAACGCUACAACUCUGGCACGUGCCAGUGGCGAAGGCGAGUUGUGUCACU------------- .((((((...(((((....)))))..(((((....)))))((-((((.........).)))))(((((((((((....))))...((....)))))))))))))))------------- ( -41.40, z-score = -1.30, R) >consensus UGGUGGCCAACACCUUGGUGGGUGCCUGGGAUCGUUCCCAGGGCGGUGGCGCCA_ACAACGCCUCCAACCUGGCCCGUGCCAGCGCCGAGGACAGUAAGUGUACAA_U___________ .(((.(((..((((......))))(((((((....))))))).....))))))........((((....(((((....)))))....))))............................ (-27.17 = -27.29 + 0.13)



| Location | 10,717,392 – 10,717,493 |
|---|---|
| Length | 101 |
| Sequences | 14 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 58.99 |
| Shannon entropy | 0.93575 |
| G+C content | 0.51517 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -14.05 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.83 |
| Mean z-score | -0.36 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10717392 101 + 27905053 AGGCGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGACAGUAAGUGUAC-AUCAGUAAGACGACCAGGAUAUGGAGAUUAUUAACUAU-------- .((((....)))).-.......(((((.(((((..((((....)(((((..(((.....)))..-.)))))...))).)))))...)))))............-------- ( -29.90, z-score = -0.63, R) >droSim1.chr3R 16773682 101 - 27517382 GGGCGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGAUAGUAAGUGUGC-ACCUUUAAGACGACCAGGAUAUGGAGAUUAUUAACCAU-------- ((..((((((((..-((....((((....(((((....)))))....))))...))..)))).)-)))..........(((.....)))..........))..-------- ( -28.20, z-score = 0.46, R) >droSec1.super_0 9902826 101 - 21120651 GGGCGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGACAGUAAGUGUGC-ACCUGUAAGACGACCAGGAUAUGGAGAUUAUUAACCAU-------- .((((....)))).-.......(((((.(((((...((((((..((((....))))...)).))-)).((.....)).)))))...)))))............-------- ( -28.60, z-score = 0.74, R) >droYak2.chr3R 13121793 99 - 28832112 GGGCGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGACAGUAAGU--AC-ACUUGUAAGACAGUCAUGUCAUGGAAAUUGUUAACUAU-------- .((((.((((((..-.....(((........)))..)))))).))))...((((((...(--((-....))).((((....))))......))))))......-------- ( -28.80, z-score = 0.15, R) >droEre2.scaffold_4770 10253583 99 + 17746568 GGGUGGUGGCGCCA-ACAACGCCUCCAACCUGGCCCGUGCCAGCGCUGAGGACAGUAAGU--AC-ACUUGUAAGACAGCCAGGGUAUGGAAAUUGUUAACUAU-------- ((((((.((((...-....)))).)).))))...(((((((...((((...(((((....--..-)).)))....))))...)))))))..............-------- ( -32.40, z-score = -0.27, R) >droAna3.scaffold_13340 13723474 98 - 23697760 GGGUGGUGGCGCCA-ACAACGCCUCCAACCUGGCUCGUGCCAGCGCUGAGGACAGUGAGU--GA-AUCUCUUAAACCACCAUUGGUUAGC-AUACUUAUCUAA-------- ((.(((.((((...-....)))).))).))((((....))))..(((((...(((((.((--(.-...........)))))))).)))))-............-------- ( -31.30, z-score = -1.01, R) >dp4.chr2 7799659 98 + 30794189 GGGCGGUGGAGCCA-ACAAUGCCUCCAACCUGGCCCGUGCCAGCGCCGAGGACAGUGAGUAUAU-AAAUCAGUAGUAAUCCCAGAUACGAGUUCACCCAG----------- (((((((((((.((-....)).))))...(((((....))))).)))).(((((.(((......-...))).).(((........)))..)))).)))..----------- ( -27.10, z-score = -0.71, R) >droPer1.super_0 1623085 98 + 11822988 GGGCGGUGGAGCCA-ACAAUGCCUCCAACCUGGCCCGUGCCAGCGCCGAGGACAGUGAGUAUAU-AAAUCAGUAGCAAUCUCACAUACGAGUUCACCCAG----------- (((((((((((.((-....)).))))...(((((....))))).)))).((((.(((((..(((-......))).....)))))......)))).)))..----------- ( -28.90, z-score = -0.97, R) >droWil1.scaffold_181130 12178581 107 - 16660200 GGGCGGUGGCGCCA-ACAAUGCAUCCAAUUUGGCUCGUGCUAGCGCCGAGGACAGUAAGUGGUU-AAGUGUUAUA--AUCUCUGAGGAAUUAUAUAAAAUUUCUGUGUUAA .((((.((((((..-.(((..........)))....)))))).))))...((((...((.((((-(.......))--))).)).(((((((......))))))).)))).. ( -26.20, z-score = 0.15, R) >droVir3.scaffold_12855 5383843 106 - 10161210 GGGCGGCGGCGCCA-AUAAUGCAUCCAACCUAGCGCGCGCCAGUGCCGAAGACAGUGAGUAUU--AAGAACCAUUUGAUCUUUGUUCUAUUGUAUA--AUCUUGGCUUUGC ..((((((((((..-.................)))).))))...(((((..(((((.((.((.--((((.(.....).)))).)).)))))))...--...)))))...)) ( -24.11, z-score = 1.55, R) >droMoj3.scaffold_6540 25002948 109 - 34148556 GGGCGGUGGCGCCA-ACAAUGCAUCCAACUUGGCCCGUGCCAGUGCCGAAGACAGUUAGUAUUCAAAUAAUCAAAAUGUCUUUGGCGCAUUGUUUGU-ACUCCUGCUUUGA ((((((..(..(.(-(((((((.....(((.(((....))))))((((((((((.....(((....))).......)))))))))))))))))).).-.)..))))))... ( -41.60, z-score = -3.42, R) >droGri2.scaffold_14906 6891161 105 - 14172833 GGGCGGUGGCGCCA-ACAACGCUUCUAAUCUGGCACGCGCCAGUGCCGAGGACAGUGAGUCAAC-AAAUCGAAGCUUAUUACGUACAGAUGCGUAAUCCUUCCUUAU---- .((((.((((((..-.....(((........)))..)))))).))))((((((.....)))...-.....((((...((((((((....)))))))).)))))))..---- ( -32.70, z-score = -1.16, R) >apiMel3.Group1 13435792 94 + 25854376 AGGAGGAGGUGUAA-ACAAUAUAAGUUAUUUGGCUGAAGCUUCUGCAACUGAUGGAUUAUGUCACAGUUAUCAAAGUUUCAAUACUUGUUAUCAA---------------- .((((.((((((..-........((((....))))(((((((..(.(((((.(((......)))))))).)..))))))).)))))).)).))..---------------- ( -17.20, z-score = 0.21, R) >triCas2.ChLG6 6402060 86 - 13544221 -AGCGUCGGCAAAACAGAACGCUACAACUCUGGCACGUGCCAGUGGCGAAGGCGAGUUGUGUCACUCGUACCAAAGUUUCAACACGU------------------------ -.((((.(...((((....((((((.....((((....))))))))))...((((((......))))))......))))...)))))------------------------ ( -25.00, z-score = -0.15, R) >consensus GGGCGGUGGCGCCA_ACAACGCCUCCAACCUGGCCCGUGCCAGCGCCGAGGACAGUAAGUGUAC_AACUGUAAAACAAUCACGGUAUGGAGAUUAUUAACUAU________ .((((.((((((....((............))....)))))).))))................................................................ (-14.05 = -13.60 + -0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:05 2011