
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,691,996 – 10,692,107 |
| Length | 111 |
| Max. P | 0.855684 |

| Location | 10,691,996 – 10,692,107 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 74.37 |
| Shannon entropy | 0.51324 |
| G+C content | 0.48593 |
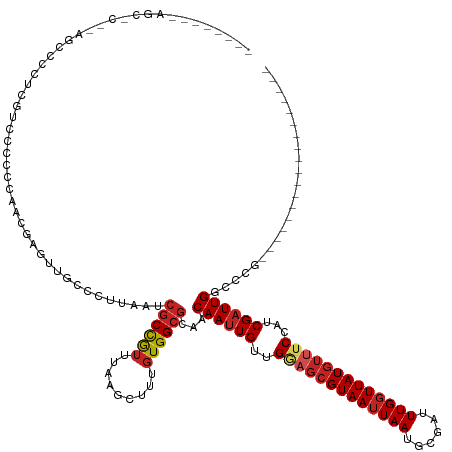
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -19.74 |
| Energy contribution | -20.26 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
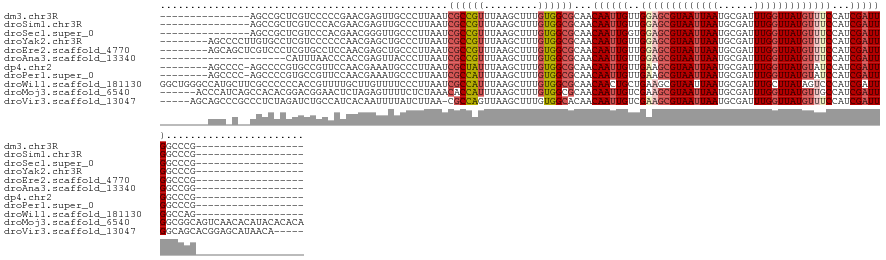
>dm3.chr3R 10691996 111 - 27905053 ---------------AGCCGCUCGUCCCCCGAACGAGUUGCCCUUAAUCGCCGUUUAAGCUUUGUGGCGCAACAAUUGUUGGAGCGUAAUUAAUGCGAUUUGGUUAUGUUUCCAUCGAUUGGCCCG------------------ ---------------.((.((((((.......)))))).))........((((((...(((....)))..)))(((((.((((((((((((((......))))))))).))))).))))))))...------------------ ( -33.80, z-score = -1.87, R) >droSim1.chr3R 16746398 111 + 27517382 ---------------AGCCGCUCGUCCCACGAACGAGUUGCCCUUAAUCGCCGUUUAAGCUUUGUGGCGCAACAAUUGUUGGAGCGUAAUUAAUGCGAUUUGGUUAUGUUUCCAUCGAUUGGCCCG------------------ ---------------.((.((((((.......)))))).))........((((((...(((....)))..)))(((((.((((((((((((((......))))))))).))))).))))))))...------------------ ( -33.80, z-score = -1.59, R) >droSec1.super_0 9877254 111 + 21120651 ---------------AGCCGCUCGUCCCACGAACGGGUUGCCCUUAAUCGCCGUUUAAGCUUUGUGGCGCAACAAUUGGUGGAGCGUAAUUAAUGCGAUUUGGUUAUGUUUCCAUCGAUUGGCCCG------------------ ---------------......(((.....))).((((((((((.(((..((.......)).))).)).))).(((((((((((((((((((((......))))))))).)))))))))))))))))------------------ ( -37.60, z-score = -1.54, R) >droYak2.chr3R 13096191 118 + 28832112 --------AGCCCCUUGUGCCUCGUCCCCCCAACGAGCUGCCCUUAAUCGCCGUUUAAGCUUUGUGGCGCAACAAUUGUUGGAGCGUAAUUAAUGCGAUUUGGUUAUGUUUCCAUCGAUUGGCCCG------------------ --------.((...(((((((.((.......((((.((...........)))))).......)).)))))))((((((.((((((((((((((......))))))))).))))).))))))))...------------------ ( -34.84, z-score = -2.36, R) >droEre2.scaffold_4770 10226793 118 - 17746568 --------AGCAGCUCGUCCCUCGUGCCUCCAACGAGCUGCCCUUAAUCGCCGUUUAAGCUUUGUGGCGCAACAAUUGUUGGAGCGUAAUUAAUGCGAUUUGGUUAUGUUUCCAUCGAUUGGCCCG------------------ --------.(((((((((..............)))))))))........((((((...(((....)))..)))(((((.((((((((((((((......))))))))).))))).))))))))...------------------ ( -39.04, z-score = -2.68, R) >droAna3.scaffold_13340 13690970 105 + 23697760 ---------------------CAUUUAACCCACCGAGUUACCCUUAAUCGCCGUUUAAGCUUUGUGGCGCAACAAUUGUUGGAGCGUAAUUAAUGCGAUUUGGUUAUGUUUCCAUCGAUUGGCCGG------------------ ---------------------...........(((.((..........(((((...........)))))...((((((.((((((((((((((......))))))))).))))).)))))))))))------------------ ( -27.60, z-score = -1.10, R) >dp4.chr2 7770990 117 - 30794189 --------AGCCCC-AGCCCCGUGCCGUUCCAACGAAAUGCCCUUAAUCGCUAUUUAAGCUUUGUGGCGCAACAAUUGUUGAAGCGUAAUUAAUGCGAUUUGGUUAUGUAUCCAUCGAUUGGCCCG------------------ --------......-.....((.((((.((((((((..(((((.(((..(((.....))).))).)).)))....))))))..((((((((((......)))))))))).......)).)))).))------------------ ( -26.90, z-score = -0.25, R) >droPer1.super_0 1594362 117 - 11822988 --------AGCCCC-AGCCCCGUGCCGUUCCAACGAAAUGCCCUUAAUCGCCAUUUAAGCUUUGUGGCGCAACAAUUGUUGAAGCGUAAUUAAUGCGAUUUGGUUAUGUAUCCAUCGAUUGGCCCG------------------ --------......-.((...((((((........((((((........).)))))........))))))..((((((..((.((((((((((......)))))))))).))...))))))))...------------------ ( -25.89, z-score = 0.10, R) >droWil1.scaffold_181130 12153504 126 + 16660200 GGCUGGGCCAUGCUUCGCCCCCCACCGUUUUGCUUGUUUUCCCUUAAUCGCCAUUUAAGCUUUGUGGCGCAACAACUGCUGAAGCGUAAUUAAUGCGAUUUGCUUAUAGUCCCAUCGAUUGGCCAG------------------ .....(((((((((((.................................(((((.........)))))(((.....))).)))))))(((..(((.((((.......)))).)))..)))))))..------------------ ( -31.40, z-score = 0.07, R) >droMoj3.scaffold_6540 29987688 138 - 34148556 ------ACCCAUCAGCCACACGGACGGAACUCUAGAGUUUUCUCUAAACACCAUUUAAGCUUUGUGGCGCAACAAUUGUCGAAGCGUAAUUAAUGCGAUUUGGUUAUGUUGCCAUCGAUUGGCGGCAGUCAACACAUACACACA ------........((((((.((..((.....(((((....)))))....)).......)).))))))......((((((..(((((((((((......)))))))))))((((.....))))))))))............... ( -34.60, z-score = -0.92, R) >droVir3.scaffold_13047 7436969 133 + 19223366 -----AGCAGCCCGCCCUCUAGAUCUGCCAUCACAAUUUUAUCUUAA-CGCCAGUUAAGCUUUGUGGCACAACAAUUGUCGAAGCGUAAUUAAUGCGAUUUGGUUAUGUUUCCAUCGAUUGGCAGCACGGAGCAUAACA----- -----.((((..(........)..))))...................-.....((((.((((((((.(....((((((..(((((((((((((......)))))))))))))...))))))...))))))))).)))).----- ( -36.50, z-score = -1.71, R) >consensus ________AGC_C__AGCCCCUCGUCCCCCCAACGAGUUGCCCUUAAUCGCCGUUUAAGCUUUGUGGCGCAACAAUUGUUGGAGCGUAAUUAAUGCGAUUUGGUUAUGUUUCCAUCGAUUGGCCCG__________________ ................................................((((((.........))))))...((((((..(((((((((((((......)))))))))))))...))))))....................... (-19.74 = -20.26 + 0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:01 2011