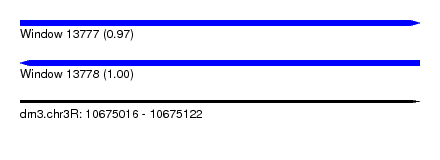
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,675,016 – 10,675,122 |
| Length | 106 |
| Max. P | 0.996375 |

| Location | 10,675,016 – 10,675,122 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Shannon entropy | 0.40031 |
| G+C content | 0.40570 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -12.49 |
| Energy contribution | -11.79 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
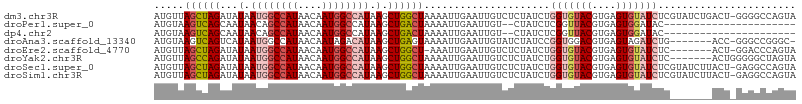
>dm3.chr3R 10675016 106 + 27905053 UACUGGCCCC-AGUCAGAUACGAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAU ..(((((...-.))))).((((.((.....)).)))).......................(((((.((...((((((((....)))))))).....)).)))))... ( -25.30, z-score = -2.65, R) >droPer1.super_0 1573986 83 + 11822988 ----------------------GUAUCCACUCACGUAACCGAGAUAG--ACAAUUCAAUUUUAGUCAGCUUAUGGCCAUUGUUAUGGCUGUUAUUGCUGACUUACAU ----------------------(((....(((........)))....--.............(((((((..((((((((....))))))))....)))))))))).. ( -23.00, z-score = -3.23, R) >dp4.chr2 7750839 83 + 30794189 ----------------------GUAUCCACUCACGUAACCGAGAUAG--ACAAUUCAAUUUUAGUCAGCUUAUGGCCAUUGUUAUGGCUGUUAUUGCUGACUUACAU ----------------------(((....(((........)))....--.............(((((((..((((((((....))))))))....)))))))))).. ( -23.00, z-score = -3.23, R) >droAna3.scaffold_13340 13674674 98 - 23697760 -GCCCGGCCC-GGU-------CAGAUCUACUCACGUCCACCGGAUAGAUACAAUUCAAUUUUACUCAGCUUAUGUCUAUUGUUAUGGCCAUUAUGACUGACUUACAU -....(((((-(((-------..(((........))).))))((((((((......................)))))))).....)))).................. ( -16.95, z-score = 0.22, R) >droEre2.scaffold_4770 10209673 98 + 17746568 UACUGGGUCC-AGU-------GAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUU-AGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAU ..((((((..-.((-------(((.....)))))..)).))))................((-(((.((...((((((((....)))))))).....)).)))))... ( -30.40, z-score = -3.91, R) >droYak2.chr3R 13078920 100 - 28832112 UACUAGCCCCCAGU-------GAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUGGCUAACAU ..(((.......((-------(((.....)))))..........))).............((((((((...((((((((....)))))))).....))))))))... ( -29.93, z-score = -4.35, R) >droSec1.super_0 9860323 106 - 21120651 UACUGGCCUC-AGUAAGAUACGAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAU ..((((.(((-.(((...)))))).(((......)))..)))).................(((((.((...((((((((....)))))))).....)).)))))... ( -24.30, z-score = -2.20, R) >droSim1.chr3R 16729455 106 - 27517382 UACUGGCCUC-AGUAAGAUACGAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAU ..((((.(((-.(((...)))))).(((......)))..)))).................(((((.((...((((((((....)))))))).....)).)))))... ( -24.30, z-score = -2.20, R) >consensus UACUGGCCCC_AGU_______GAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAU ..............................................................(((.((...((((((((....)))))))).....)).)))..... (-12.49 = -11.79 + -0.70)

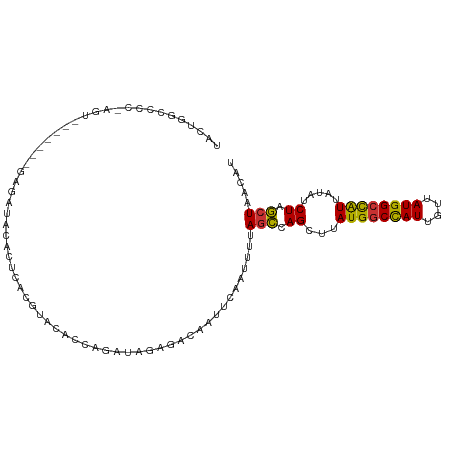
| Location | 10,675,016 – 10,675,122 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Shannon entropy | 0.40031 |
| G+C content | 0.40570 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10675016 106 - 27905053 AUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUCGUAUCUGACU-GGGGCCAGUA ...((((((((...(.((((((((....)))))))).).))))))))......((((.(((((..(((.(((((.((.....)).))))))))..)-)))).)))). ( -35.40, z-score = -3.00, R) >droPer1.super_0 1573986 83 - 11822988 AUGUAAGUCAGCAAUAACAGCCAUAACAAUGGCCAUAAGCUGACUAAAAUUGAAUUGU--CUAUCUCGGUUACGUGAGUGGAUAC---------------------- .....(((((((.......(((((....))))).....)))))))..........(((--(((((.((....)).)).)))))).---------------------- ( -23.20, z-score = -2.90, R) >dp4.chr2 7750839 83 - 30794189 AUGUAAGUCAGCAAUAACAGCCAUAACAAUGGCCAUAAGCUGACUAAAAUUGAAUUGU--CUAUCUCGGUUACGUGAGUGGAUAC---------------------- .....(((((((.......(((((....))))).....)))))))..........(((--(((((.((....)).)).)))))).---------------------- ( -23.20, z-score = -2.90, R) >droAna3.scaffold_13340 13674674 98 + 23697760 AUGUAAGUCAGUCAUAAUGGCCAUAACAAUAGACAUAAGCUGAGUAAAAUUGAAUUGUAUCUAUCCGGUGGACGUGAGUAGAUCUG-------ACC-GGGCCGGGC- ..((..(((((((.......((((....(((((.((((..(((......)))..)))).)))))...))))((....)).)).)))-------)).-..)).....- ( -19.60, z-score = 0.79, R) >droEre2.scaffold_4770 10209673 98 - 17746568 AUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCU-AAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUC-------ACU-GGACCCAGUA ...((((((((...(.((((((((....)))))))).).))))))-))................((((.((..(((((.....)))-------)).-..)))))).. ( -34.50, z-score = -3.70, R) >droYak2.chr3R 13078920 100 + 28832112 AUGUUAGCCAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUC-------ACUGGGGGCUAGUA ...((((((((...(.((((((((....)))))))).).)))))))).........(((((((...(((((((....)))))))..-------..)))))))..... ( -38.70, z-score = -4.26, R) >droSec1.super_0 9860323 106 + 21120651 AUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUCGUAUCUUACU-GAGGCCAGUA ...((((((((...(.((((((((....)))))))).).)))))))).((((....(((((.....((.(((((.((.....)).)))))))....-))))))))). ( -34.40, z-score = -3.19, R) >droSim1.chr3R 16729455 106 + 27517382 AUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUCGUAUCUUACU-GAGGCCAGUA ...((((((((...(.((((((((....)))))))).).)))))))).((((....(((((.....((.(((((.((.....)).)))))))....-))))))))). ( -34.40, z-score = -3.19, R) >consensus AUGUUAGCCAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUC_______ACU_GGGGCCAGUA .....((((((...(.((((((((....)))))))).).)))))).....................(((((((....)))))))....................... (-20.73 = -21.32 + 0.59)
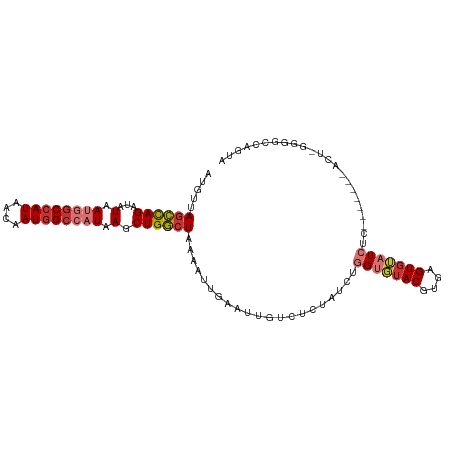
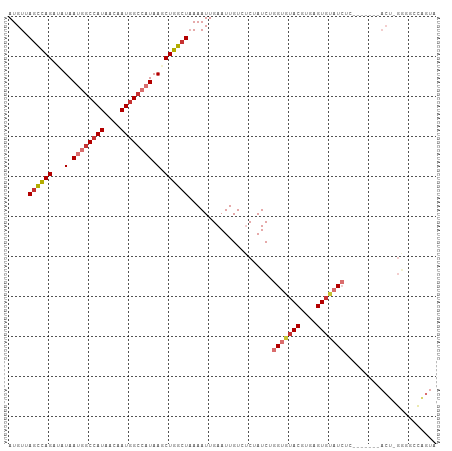
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:57 2011