| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,667,981 – 10,668,082 |
| Length | 101 |
| Max. P | 0.996252 |

| Location | 10,667,981 – 10,668,082 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 63.44 |
| Shannon entropy | 0.67127 |
| G+C content | 0.33168 |
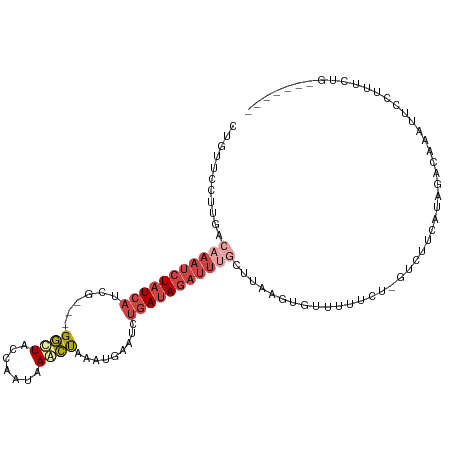
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -7.16 |
| Energy contribution | -9.30 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.996012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
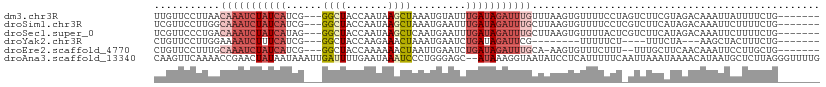
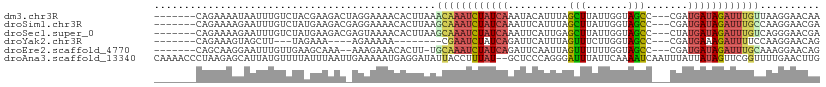
>dm3.chr3R 10667981 101 + 27905053 UUGUUCCUUAACAAAUCUAUCAUCG---GGCUACCAAUAAGCUAAAUGUAUUUGAUAGAUUUGUUUAAGUGUUUUCCUAGUCUUCGUAGACAAAUUAUUUUCUG------- ......((((((((((((((((..(---(....)).....((.....))...))))))))))).)))))..........((((....)))).............------- ( -21.90, z-score = -3.22, R) >droSim1.chr3R 16720750 101 - 27517382 UCGUUCCUUGGCAAAUCUAUCAUCG---GGCUACCAAUAAGCUAAAUGAAUUUGAUAGAUUUGCUUAAGUGUUUUCCUCGUCUUCAUAGACAAAUUCUUUUCUG------- ......(((((((((((((((((((---((((.......))))...)))...)))))))))))).))))..........((((....)))).............------- ( -24.50, z-score = -3.46, R) >droSec1.super_0 9853505 101 - 21120651 UCGUUCCCUGACAAAUCUAUCAUAG---GGCUACCAAUAAGCUCAAUGAAUUUGAUAGAUUUGCUUAAGUGUUUUACUCGUCUUCAUAGACAAAUUCUUUUCUG------- ........((((((((((((((..(---((((.......)))))........))))))))))).)))............((((....)))).............------- ( -19.60, z-score = -1.77, R) >droYak2.chr3R 13071609 86 - 28832112 CUGUUCCUUGGAAAAUCUUUCAUCG---GGCUACCAAGAAACUAAAUGAAUCUGAUAGAUUCG--------UUUUUCU----UUUCUA---AAGCUACUUUCUG------- ..((((..(((((....)))))..)---))).....(((((..(((((((((.....))))))--------)))....----))))).---.............------- ( -15.90, z-score = -1.57, R) >droEre2.scaffold_4770 10202523 98 + 17746568 CUGUUCCUUUGCAAAUCUAUCAUCG---GGCUACCAAAAAACUAAUUGAAUCUGAUAGAUUUGCA-AAGUGUUUCUUU--UUUGCUUCAACAAAUUCCUUGCUG------- ......((((((((((((((((..(---(....))......(.....)....)))))))))))))-))).((......--((((......))))......))..------- ( -23.00, z-score = -3.71, R) >droAna3.scaffold_13340 13669476 109 - 23697760 CAAGUUCAAAACCGAACUAUAAUAAAUUGAUUUUGAAUAAAUCCCUGGGAGC--AUAAAGGUAAUAUCCUCAUUUUUCAAUUAAAUAAAACAUAAUGCUCUUAGGGUUUUG ...((((((((.(((...........))).))))))))(((.((((((((((--((..(((......))).((((.......))))........)))))))))))).))). ( -25.50, z-score = -3.59, R) >consensus CUGUUCCUUGACAAAUCUAUCAUCG___GGCUACCAAUAAACUAAAUGAAUCUGAUAGAUUUGCUUAAGUGUUUUUCU_GUCUUCAUAGACAAAUUCCUUUCUG_______ ...........((((((((((((((...((((.......))))...)))...)))))))))))................................................ ( -7.16 = -9.30 + 2.14)
| Location | 10,667,981 – 10,668,082 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 63.44 |
| Shannon entropy | 0.67127 |
| G+C content | 0.33168 |
| Mean single sequence MFE | -22.41 |
| Consensus MFE | -9.26 |
| Energy contribution | -9.71 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.996252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
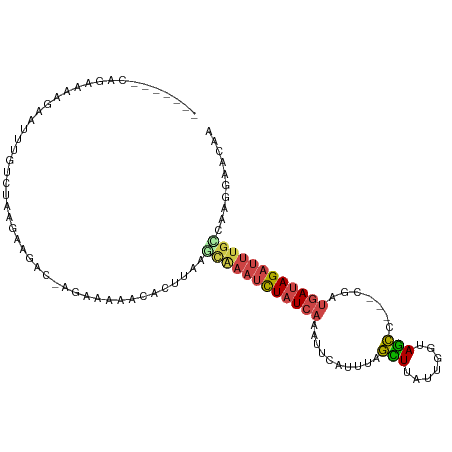
>dm3.chr3R 10667981 101 - 27905053 -------CAGAAAAUAAUUUGUCUACGAAGACUAGGAAAACACUUAAACAAAUCUAUCAAAUACAUUUAGCUUAUUGGUAGCC---CGAUGAUAGAUUUGUUAAGGAACAA -------.............((((....))))..........(((.(((((((((((((...............((((....)---)))))))))))))))).)))..... ( -19.96, z-score = -2.49, R) >droSim1.chr3R 16720750 101 + 27517382 -------CAGAAAAGAAUUUGUCUAUGAAGACGAGGAAAACACUUAAGCAAAUCUAUCAAAUUCAUUUAGCUUAUUGGUAGCC---CGAUGAUAGAUUUGCCAAGGAACGA -------..........(((((((....))))))).......(((..((((((((((((...............((((....)---)))))))))))))))..)))..... ( -22.76, z-score = -2.28, R) >droSec1.super_0 9853505 101 + 21120651 -------CAGAAAAGAAUUUGUCUAUGAAGACGAGUAAAACACUUAAGCAAAUCUAUCAAAUUCAUUGAGCUUAUUGGUAGCC---CUAUGAUAGAUUUGUCAGGGAACGA -------.........((((((((....))))))))......(((..((((((((((((.....((..(.....)..))....---...))))))))))))..)))..... ( -22.30, z-score = -1.70, R) >droYak2.chr3R 13071609 86 + 28832112 -------CAGAAAGUAGCUU---UAGAAA----AGAAAAA--------CGAAUCUAUCAGAUUCAUUUAGUUUCUUGGUAGCC---CGAUGAAAGAUUUUCCAAGGAACAG -------.............---......----.......--------.(((((.....))))).....(((((((((.....---(.......).....))))))))).. ( -15.60, z-score = -0.95, R) >droEre2.scaffold_4770 10202523 98 - 17746568 -------CAGCAAGGAAUUUGUUGAAGCAAA--AAAGAAACACUU-UGCAAAUCUAUCAGAUUCAAUUAGUUUUUUGGUAGCC---CGAUGAUAGAUUUGCAAAGGAACAG -------(((((((...))))))).......--.........(((-(((((((((((((......(((((....)))))....---...))))))))))))))))...... ( -26.72, z-score = -3.56, R) >droAna3.scaffold_13340 13669476 109 + 23697760 CAAAACCCUAAGAGCAUUAUGUUUUAUUUAAUUGAAAAAUGAGGAUAUUACCUUUAU--GCUCCCAGGGAUUUAUUCAAAAUCAAUUUAUUAUAGUUCGGUUUUGAACUUG .....((((..((((((...((..(((((.(((....)))..)))))..))....))--))))..)))).....(((((((((((((......)))).))))))))).... ( -27.10, z-score = -3.54, R) >consensus _______CAGAAAAGAAUUUGUCUAAGAAGAC_AGAAAAACACUUAAGCAAAUCUAUCAAAUUCAUUUAGCUUAUUGGUAGCC___CGAUGAUAGAUUUGCCAAGGAACAA ...............................................((((((((((((..........(((.......))).......)))))))))))).......... ( -9.26 = -9.71 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:54 2011