| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,612,622 – 10,612,720 |
| Length | 98 |
| Max. P | 0.590130 |

| Location | 10,612,622 – 10,612,720 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.13 |
| Shannon entropy | 0.49306 |
| G+C content | 0.37431 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -13.28 |
| Energy contribution | -12.90 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.590130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
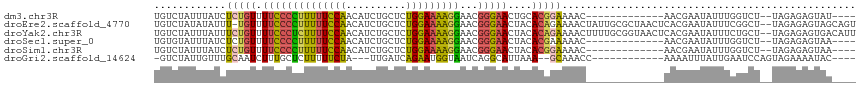
>dm3.chr3R 10612622 98 - 27905053 UGUCUAUUUAUCUCUGUUUUCCCCUUUUUCCAACAUCUGCUCUGGAAAAGGAACGGGAACUGCACGGAAAAC-------------AACGAAUAUUUGGUCU--UAGAGAGUAU---- ..........((((((((((((.(((((((((..........)))))))))...(....).....)))))))-------------..(((....)))....--.)))))....---- ( -21.80, z-score = -0.64, R) >droEre2.scaffold_4770 10146924 114 - 17746568 UGUCUAUAUAUUU-UGUUUUCCCCUUUUUCCAACAUCUGCUCUGGAAAAGGAACGGGAACUACACAGAAAACUAUUGCGCUAACUCACGAAUAUUUCGGCU--UAGAGAGUAGCAGU ..........(((-(((.((((((((((((((..........)))))))))...)))))....)))))).........((((.(((.(((.....)))...--..)))..))))... ( -24.40, z-score = -0.81, R) >droYak2.chr3R 13015319 115 + 28832112 UGUCUAUUUAUUUCUGUUUUCCCUCUUUUCCAACAUCUGCUCUGGAAAAGGAACGGGAACUACACAGAAAACUUUUGCGGUAACUCACGAAUAUUUCUGCU--UAGAGAGUGACAUU ((((......(((((((.((((((((((((((..........)))))))))...)))))....)))))))((((((((((................)))).--..)))))))))).. ( -29.89, z-score = -2.78, R) >droSec1.super_0 9798642 98 + 21120651 UGUGUAUUUAUCUCUGUUUUCCCCUUUUUCCAACAUCUGCUCUGGAAAAGGAACGGGAACUACACGAAAAAC-------------AACGAAUAUUUGGUCU--UAGAGAGUAA---- ..........((((((..((((((((((((((..........)))))))))...))))).....((......-------------..))............--))))))....---- ( -19.90, z-score = -0.37, R) >droSim1.chr3R 16669352 98 + 27517382 UGUCUAUUUAUCUCUGUUUUCCCCUUUUUCCAACAUCUGCUCUGGAAAAGGAACGGGAACUACACGGAAAAC-------------AACGAAUAUUUGGUCU--UAGAGAGUAA---- ..........((((((((((((.(((((((((..........)))))))))...(....).....)))))))-------------..(((....)))....--.)))))....---- ( -21.80, z-score = -0.78, R) >droGri2.scaffold_14624 1774389 95 - 4233967 -GUCUAUUGUUUGCAAUCUUUGCUCUUUUUCUA---UUGAUCAGAAUGGUAAUCAGGCAUUAAA--GCAAACC------------AAAAUUUAUUGAAUCCAGUAGAAAAUAC---- -.......((((((......((((......(((---((......)))))......)))).....--)))))).------------....(((((((....)))))))......---- ( -16.30, z-score = -0.77, R) >consensus UGUCUAUUUAUCUCUGUUUUCCCCUUUUUCCAACAUCUGCUCUGGAAAAGGAACGGGAACUACACGGAAAAC_____________AACGAAUAUUUGGUCU__UAGAGAGUAA____ ............(((((.((((((((((((((..........)))))))))...)))))....)))))................................................. (-13.28 = -12.90 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:49 2011