
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,564,639 – 10,564,729 |
| Length | 90 |
| Max. P | 0.648876 |

| Location | 10,564,639 – 10,564,729 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 72.90 |
| Shannon entropy | 0.55786 |
| G+C content | 0.34293 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -7.62 |
| Energy contribution | -7.64 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10564639 90 - 27905053 GUCAUGGCCAUUGAGUUUUCCUCGUUUUUCGUAGUUUUCUAUCCAUUAUAACAUU--UUAAUGCCU--GCCUAAAUGAAAACGAAAUGGCAAGC ......((((((..((((((..........((((....)))).(((((.......--.)))))...--........))))))..)))))).... ( -18.20, z-score = -1.77, R) >droGri2.scaffold_14624 1716273 72 - 4233967 GUCAUGGCCAUUGAAUUUUU-------------------UCCUUAUUAUAAACUU--AUGUUCAAUU-GAUUAAAUGAAAAUGAAAUGCUCCUA .....((.((((..((((((-------------------.....((((..(((..--..)))....)-))).....))))))..))))..)).. ( -7.50, z-score = -0.09, R) >droVir3.scaffold_13047 11642498 88 - 19223366 GUCAUGGCCAUUGAAUUUUUC----CUUUGGUGCUGCUGUUUUUGUUAUAAACUUUGAUACGUAUUUAAAUGAAAUGAAAACGAAAUGCUAC-- ......((((..(........----)..))))((...((((((..((.((...(((((.......))))))).))..))))))....))...-- ( -10.30, z-score = 0.65, R) >droAna3.scaffold_13340 13567013 89 + 23697760 GUCAUGGCCAUUGAGUUUUCCUCUUUAUUUUUCCCCAUUU-GCCAUUAUAAAAUU--AUUUUCCCC--AACAACAUGAAAACGAAUUUGUGAGU ....((((....(((.....))).................-))))(((((((.(.--.(((((...--........)))))..).))))))).. ( -8.50, z-score = 0.41, R) >droEre2.scaffold_4770 10098678 86 - 17746568 GUCAUGGCCAUUGAGUUUUCCUCGUUUUUCGUAGUUU----UCCAUUAUAACAUU--UUAAUGCCU--GUCUAAAUGAAAACGAAAUGGCAAGC ......(((((((((.....)))((((((..(((...----..(((((.......--.)))))...--..)))...))))))..)))))).... ( -18.10, z-score = -1.90, R) >droYak2.chr3R 12965710 90 + 28832112 GUCAUGGCCAUUGAGUUUUCCUCAUUUUUCGUAGUUUUCUUUCCAUUAUAACAUU--UUAAUGCCU--GCCUAAAUGAAAACGAAAUGGCAAGC ......((((((..((((...((((((...((((.........(((((.......--.))))).))--))..))))))))))..)))))).... ( -18.40, z-score = -1.94, R) >droSec1.super_0 9751648 89 + 21120651 GUCAUGGCCAUUGAGUUUUCCUCG-UUUUCGUAGUUUUCUAUCCAUUAUAACAUU--UUAAUGCCU--GCCUAAAUGAAAACGAAAUGGCAAGC ......((((((..((((((...(-(((..((((.........(((((.......--.))))).))--))..))))))))))..)))))).... ( -18.90, z-score = -2.02, R) >droSim1.chr3R 16620921 89 + 27517382 GUCAUGGCCAUUGAGUUUUCCUCG-UUUUCGUAGUUUUCUAUCCAUUAUAACAUU--UUAAUGCCU--GCCUAAAUGAAAACGAAAUGGCAAGC ......((((((..((((((...(-(((..((((.........(((((.......--.))))).))--))..))))))))))..)))))).... ( -18.90, z-score = -2.02, R) >consensus GUCAUGGCCAUUGAGUUUUCCUCG_UUUUCGUAGUUUUCUAUCCAUUAUAACAUU__UUAAUGCCU__GCCUAAAUGAAAACGAAAUGGCAAGC ......((((((..((((((........................................................))))))..)))))).... ( -7.62 = -7.64 + 0.02)
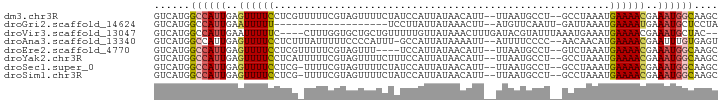

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:44 2011