| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,544,025 – 10,544,125 |
| Length | 100 |
| Max. P | 0.647895 |

| Location | 10,544,025 – 10,544,125 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.45 |
| Shannon entropy | 0.33588 |
| G+C content | 0.38686 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.50 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
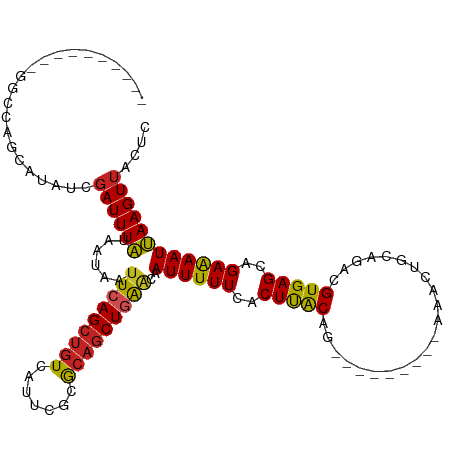
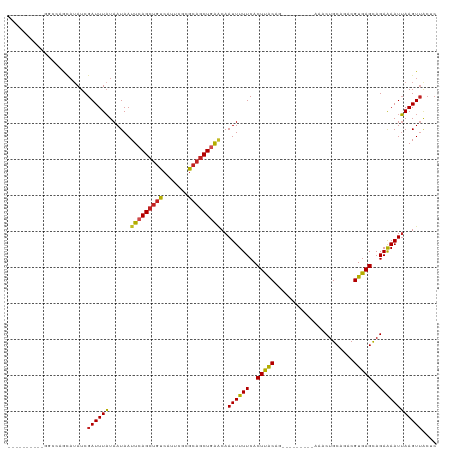
>dm3.chr3R 10544025 100 - 27905053 ----------GGCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCGCGCAGCUGAACAAUUUUUCACUUACAG---------AAACUGCAGACGUGAGCAGAAAAUUAAGUUACUC ----------....(((.................(((((((((.......))))))))).(((((((..(((((..---------...........)))))..)))))))..))).... ( -19.42, z-score = -0.53, R) >droPer1.super_0 1418046 101 - 11822988 ----------GUCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCGCGCAGCUAAACAAUUUUUCACUUACAG--------AAAACUGCAAACGUGAGUAGAAAAUUAAGUUACUC ----------....(((....................((((((.......))))))....((((((..((((((..--------............))))))..))))))..))).... ( -16.44, z-score = -0.83, R) >dp4.chr2 7583894 101 - 30794189 ----------GUCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCGCGCAGCUAAACAAUUUUUCACUUACAG--------AAAACUGCAAACGUGAGCAGAAAAUUAAGUUACUC ----------............((((((.........((((((.......)))))).......(((((.......)--------))))((((........)))).....)))))).... ( -14.20, z-score = 0.14, R) >droSim1.chr3R 16599812 100 + 27517382 ----------GGCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCGCGCAGCUGAACAAUUUUUCACUUACAG---------AAACUGCAGACGUGAGCAGAAAAUUAAGUUACUC ----------....(((.................(((((((((.......))))))))).(((((((..(((((..---------...........)))))..)))))))..))).... ( -19.42, z-score = -0.53, R) >droSec1.super_0 9731340 100 + 21120651 ----------GGCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCGCGCAGCUGAACAAUUUUUCACUUACAG---------AAACUGCAGACGUGAGCAGAAAAUUAAGUUACUC ----------....(((.................(((((((((.......))))))))).(((((((..(((((..---------...........)))))..)))))))..))).... ( -19.42, z-score = -0.53, R) >droYak2.chr3R 12944689 100 + 28832112 ----------GGCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCGCGCAGCUGAACAAUUUUUCACUUACAG---------AAACUGCAGACGUGAGCAGAAAAUUAAGUUACUC ----------....(((.................(((((((((.......))))))))).(((((((..(((((..---------...........)))))..)))))))..))).... ( -19.42, z-score = -0.53, R) >droEre2.scaffold_4770 10078030 100 - 17746568 ----------GGCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCGCGCAGCUGAACAAUUUUUCACUUACAG---------AAACUGCAGACGUGAGCAGAAAAUUAAGUUACUC ----------....(((.................(((((((((.......))))))))).(((((((..(((((..---------...........)))))..)))))))..))).... ( -19.42, z-score = -0.53, R) >droWil1.scaffold_181089 9674099 87 + 12369635 -------------------AUCGAUUUAUAAUAAUUCAGCUGUCAUUUGCACAGCUGAACAAUUUUUCACUCACAG---------AAACUGCAAACGUGAGCAGAAAAUUAAGUU---- -------------------...............((((((((((....).))))))))).(((((((..(((((..---------...........)))))..))))))).....---- ( -21.02, z-score = -2.86, R) >droVir3.scaffold_12855 8527104 103 + 10161210 ----------------CAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCCCGCAGCUGGCCAAUUUUUCACUCGCCUCGCACUGCAAAACUGCAAACGUGAGCAGAAAAUUAAGUUGCCC ----------------.....(((((((.......((((((((.......))))))))............((..(((((..((((....))))...)))))..))....)))))))... ( -22.90, z-score = -1.66, R) >droMoj3.scaffold_6540 6787910 109 + 34148556 ----------GUCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUCCCGGCAGCUGAGCAAUUUUUCACUCACUUUGCGCUGCAGAACUGCAGACGUGAGCAGAGAAUUAAGUGGCCC ----------..((((((((......)))......(((((((((.....)))))))))))(((((((..(((((......(((((....)))))..)))))..)))))))...)))... ( -33.70, z-score = -2.41, R) >droGri2.scaffold_14624 58820 118 + 4233967 GUCUGAAAUAUAUAUAUAUAUCGAUUUUUAAUAAUUCAGGCUUCAUUCUCACAGCUGGGCAAUUUUUCACUCGCACUUCAGCA-AAAACUGCAAACGUGAGCAGAAAAUGAAGUUGCCC (((((((.(((...................))).)))))))...............((((((((((......((......)).-....((((........)))).....)))))))))) ( -25.71, z-score = -1.89, R) >consensus __________GGCCAGCAUAUCGAUUUAUAAUAAUUCAGCUGUCAUUCGCGCAGCUGAACAAUUUUUCACUUACAG_________AAACUGCAGACGUGAGCAGAAAAUUAAGUUACUC ......................((((((......(((((((((.......)))))))))..((((((..(((((......................)))))..)))))))))))).... (-16.92 = -16.50 + -0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:42 2011