
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,543,009 – 10,543,100 |
| Length | 91 |
| Max. P | 0.766874 |

| Location | 10,543,009 – 10,543,100 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.51540 |
| G+C content | 0.49334 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -11.74 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
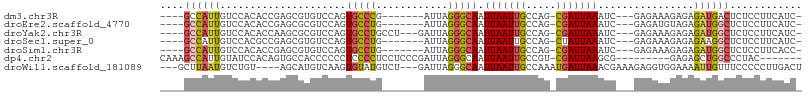
>dm3.chr3R 10543009 91 + 27905053 ----GCCAUUGUCCACACCGAGCGUGUCCAGUGCCCG-------AUUAGGGCAAUUAAUUGCCAG-CGAUUAAAUC---GAGAAAGAGAGAUGACUCUCCUUCAUC- ----...((((..(((.......)))..))))((((.-------....))))..(((((((....-)))))))...---..(((.(((((....))))).)))...- ( -26.70, z-score = -2.72, R) >droEre2.scaffold_4770 10076920 91 + 17746568 ----GCCAUUGUCCACACCGAGCGCGUCCAGUGCCUG-------AUUAGGGCAAUUAAUUGCCAG-CGAUUAAAUC---GAGAUGUAGAGAUGGCUCUCCUUCAUC- ----((((((.((.(((((((((((.....)))).((-------(((..(((((....)))))..-.)))))..))---).).))).))))))))...........- ( -26.00, z-score = -1.29, R) >droYak2.chr3R 12943615 95 - 28832112 ----GCCAUUGUCCACACCAAGCGCGUCCAGUGCCUGCCU---GAUUAGGGCAAUUAAUUGCCAG-CGAUUAAAUC---GAGAAAGAGAGAUGGCUCUCCUUCAUC- ----(((...(((.....((.((((.....)))).))...---)))...)))..(((((((....-)))))))...---..(((.(((((....))))).)))...- ( -26.00, z-score = -1.08, R) >droSec1.super_0 9730327 91 - 21120651 ----GCCAUUGUCCACGCCGAGCGUGUCCAGUGCCUG-------AUUAGGGCAAUUAAUUGCCAG-CUAUUAAAUC---GAGAAAGAGAGAAGGCUCUCCUUCAUC- ----(((((((..(((((...)))))..)))))....-------.....(((((....))))).)-).........---..(((.(((((....))))).)))...- ( -28.20, z-score = -2.26, R) >droSim1.chr3R 16598796 91 - 27517382 ----GCCAUUGUCCACACCGAGCGUGUCCAGUGCCUG-------AUUAGGGCAAUUAAUUGCCAG-CGAUUAAAUC---GAGAAAGAGAGAUGGCUCUCCUUCACC- ----(.(((((..(((.......)))..))))).)((-------(((..(((((....)))))..-.)))))....---..(((.(((((....))))).)))...- ( -24.30, z-score = -1.18, R) >dp4.chr2 7583165 90 + 30794189 CAAAGCCAUUGUAUCCACAGUGCCACCCCCCUCCCCUCCUCCCGAUUAGGGCAAUUAAUUGCCGU-CGAUUAAGCG---------GAGAGCUGGCCCUAC------- ....((((((((....))))).........((((.((.....((((...(((((....)))))))-))....)).)---------)))....))).....------- ( -20.40, z-score = -0.53, R) >droWil1.scaffold_181089 9673070 97 - 12369635 ---GCUUAAUGUCUGU----AGCAUGUCAAGUGUAUGUCU---GAUUAGGGCAAUUAAUUGCCAAAUGAUUAAACGAAAGAGGUGGAAAAUUGUUUCCCCCUUGACU ---(((..........----)))..((((((....(((.(---(((((.(((((....)))))...)))))).))).....((.(((((....))))))))))))). ( -26.10, z-score = -2.64, R) >consensus ____GCCAUUGUCCACACCGAGCGUGUCCAGUGCCUG_______AUUAGGGCAAUUAAUUGCCAG_CGAUUAAAUC___GAGAAAGAGAGAUGGCUCUCCUUCAUC_ ....((((((........((.((((.....)))).))............(((((....)))))..........................))))))............ (-11.74 = -11.87 + 0.13)
| Location | 10,543,009 – 10,543,100 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.51540 |
| G+C content | 0.49334 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -14.25 |
| Energy contribution | -14.81 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.751318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
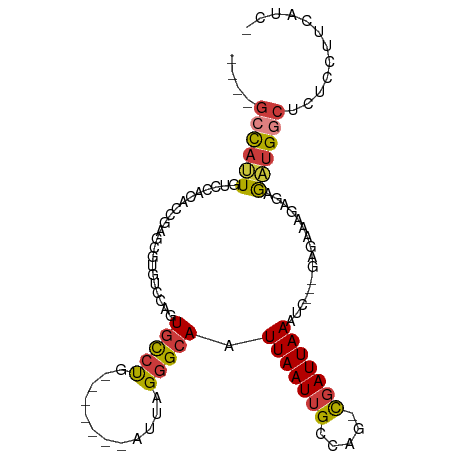
>dm3.chr3R 10543009 91 - 27905053 -GAUGAAGGAGAGUCAUCUCUCUUUCUC---GAUUUAAUCG-CUGGCAAUUAAUUGCCCUAAU-------CGGGCACUGGACACGCUCGGUGUGGACAAUGGC---- -((.(((((((((....)))))))))))---.........(-((..........(((((....-------.))))).((..(((((...)))))..))..)))---- ( -30.60, z-score = -2.52, R) >droEre2.scaffold_4770 10076920 91 - 17746568 -GAUGAAGGAGAGCCAUCUCUACAUCUC---GAUUUAAUCG-CUGGCAAUUAAUUGCCCUAAU-------CAGGCACUGGACGCGCUCGGUGUGGACAAUGGC---- -...........(((((.((((((((..---(((...)))(-(.(((((....)))))....(-------(((...))))..))....))))))))..)))))---- ( -23.40, z-score = 0.09, R) >droYak2.chr3R 12943615 95 + 28832112 -GAUGAAGGAGAGCCAUCUCUCUUUCUC---GAUUUAAUCG-CUGGCAAUUAAUUGCCCUAAUC---AGGCAGGCACUGGACGCGCUUGGUGUGGACAAUGGC---- -((.(((((((((....)))))))))))---.........(-(((((((....)))))......---.))).(.((.((..(((((...)))))..)).)).)---- ( -28.61, z-score = -0.69, R) >droSec1.super_0 9730327 91 + 21120651 -GAUGAAGGAGAGCCUUCUCUCUUUCUC---GAUUUAAUAG-CUGGCAAUUAAUUGCCCUAAU-------CAGGCACUGGACACGCUCGGCGUGGACAAUGGC---- -((.(((((((((....)))))))))))---(((....(((-..(((((....))))))))))-------).(.((.((..(((((...)))))..)).)).)---- ( -30.00, z-score = -2.23, R) >droSim1.chr3R 16598796 91 + 27517382 -GGUGAAGGAGAGCCAUCUCUCUUUCUC---GAUUUAAUCG-CUGGCAAUUAAUUGCCCUAAU-------CAGGCACUGGACACGCUCGGUGUGGACAAUGGC---- -.(.(((((((((....))))))))).)---((((......-..(((((....)))))..)))-------).(.((.((..(((((...)))))..)).)).)---- ( -27.10, z-score = -1.04, R) >dp4.chr2 7583165 90 - 30794189 -------GUAGGGCCAGCUCUC---------CGCUUAAUCG-ACGGCAAUUAAUUGCCCUAAUCGGGAGGAGGGGAGGGGGGUGGCACUGUGGAUACAAUGGCUUUG -------.((((((((.(((((---------(.(((..(((-(.(((((....)))))....))))...))).))))))..(((.(......).)))..)))))))) ( -31.70, z-score = -1.09, R) >droWil1.scaffold_181089 9673070 97 + 12369635 AGUCAAGGGGGAAACAAUUUUCCACCUCUUUCGUUUAAUCAUUUGGCAAUUAAUUGCCCUAAUC---AGACAUACACUUGACAUGCU----ACAGACAUUAAGC--- .(((((((((((((....))))).))......((((........(((((....)))))......---)))).....)))))).....----.............--- ( -20.84, z-score = -2.18, R) >consensus _GAUGAAGGAGAGCCAUCUCUCUUUCUC___GAUUUAAUCG_CUGGCAAUUAAUUGCCCUAAU_______CAGGCACUGGACACGCUCGGUGUGGACAAUGGC____ ....((.((((((....)))))).))..................(((((....)))))..............(.((.((..(((((...)))))..)).)).).... (-14.25 = -14.81 + 0.57)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:41 2011