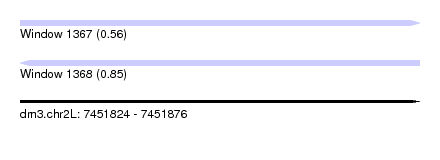
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,451,824 – 7,451,876 |
| Length | 52 |
| Max. P | 0.853419 |

| Location | 7,451,824 – 7,451,876 |
|---|---|
| Length | 52 |
| Sequences | 7 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 74.39 |
| Shannon entropy | 0.46576 |
| G+C content | 0.42339 |
| Mean single sequence MFE | -12.18 |
| Consensus MFE | -7.56 |
| Energy contribution | -7.70 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559525 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
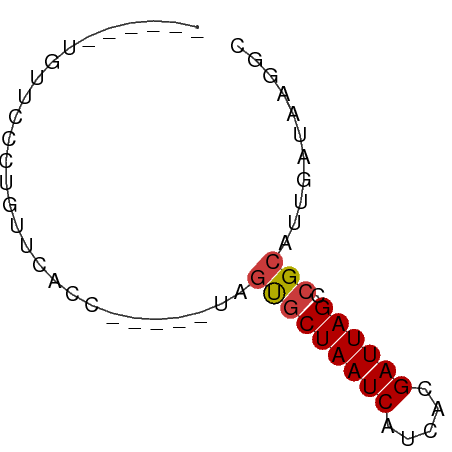
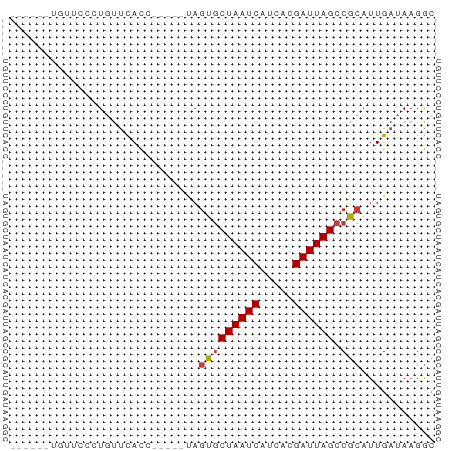
>dm3.chr2L 7451824 52 + 23011544 GCCUUAUCAAUGCGGCUAAUCGUGAUGAUUAGCACUG-----GGUGAACAGCGAACA------ ((.(((((......(((((((.....)))))))....-----)))))...)).....------ ( -12.30, z-score = -0.59, R) >droYak2.chr2L 16878051 57 - 22324452 GCCUUAUCAAUGCGGCUAAUCGUGAUGAUUAGCACUAACAUAGGUGAACACGGCACA------ ((((((((.(((..(((((((.....))))))).....))).)))))....)))...------ ( -15.70, z-score = -2.12, R) >droSec1.super_3 2968494 52 + 7220098 GCCUUAUCAAUGCGGCUAAUCGUGAUGAUUAGCACUA-----GGUGAACAGCGAACA------ ((....(((.....(((((((.....)))))))....-----..)))...)).....------ ( -11.30, z-score = -0.50, R) >droSim1.chr2L 7243745 52 + 22036055 GCCUUAUCAAUGCGGCUAAUCGUGAUGAUUAGCACUA-----GGUGAACAGCGAACA------ ((....(((.....(((((((.....)))))))....-----..)))...)).....------ ( -11.30, z-score = -0.50, R) >droVir3.scaffold_12963 4820263 55 - 20206255 GCCAUAUCAAUGCGGCUAAUCGAAGUGAUUAGCGCUA-----AGUGAACAAAUACUUACA--- ......(((..(((.((((((.....)))))))))..-----..))).............--- ( -11.90, z-score = -1.46, R) >droMoj3.scaffold_6500 13990195 55 - 32352404 GUCUUAUCAAUGCGGCUAAUCAUUCUGAUUAGAAAUA---UAUAUAUAAAUGCAUGUA----- .........(((((.((((((.....)))))).....---..........)))))...----- ( -7.99, z-score = -0.36, R) >droGri2.scaffold_15126 5037947 58 + 8399593 GCCUUAUCAAUGCGGCUAAUCGAUGUGAUUAGCGCUA-----AGUGAACAUGCACUUAAUCCA ...........(((.((((((.....)))))))))((-----((((......))))))..... ( -14.80, z-score = -1.71, R) >consensus GCCUUAUCAAUGCGGCUAAUCGUGAUGAUUAGCACUA_____GGUGAACAGCGAACA______ ..............(((((((.....))))))).............................. ( -7.56 = -7.70 + 0.14)

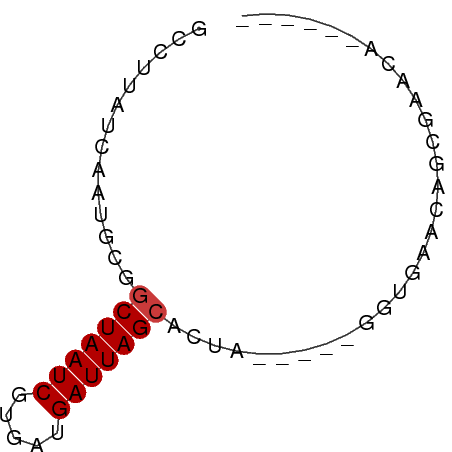
| Location | 7,451,824 – 7,451,876 |
|---|---|
| Length | 52 |
| Sequences | 7 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 74.39 |
| Shannon entropy | 0.46576 |
| G+C content | 0.42339 |
| Mean single sequence MFE | -12.51 |
| Consensus MFE | -8.22 |
| Energy contribution | -8.30 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853419 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
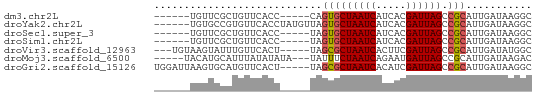
>dm3.chr2L 7451824 52 - 23011544 ------UGUUCGCUGUUCACC-----CAGUGCUAAUCAUCACGAUUAGCCGCAUUGAUAAGGC ------.....(((..(((..-----..(.(((((((.....))))))))....)))...))) ( -13.10, z-score = -1.61, R) >droYak2.chr2L 16878051 57 + 22324452 ------UGUGCCGUGUUCACCUAUGUUAGUGCUAAUCAUCACGAUUAGCCGCAUUGAUAAGGC ------...(((....(((...((((..(.(((((((.....)))))))))))))))...))) ( -14.40, z-score = -1.24, R) >droSec1.super_3 2968494 52 - 7220098 ------UGUUCGCUGUUCACC-----UAGUGCUAAUCAUCACGAUUAGCCGCAUUGAUAAGGC ------.....(((..(((..-----..(.(((((((.....))))))))....)))...))) ( -13.10, z-score = -1.53, R) >droSim1.chr2L 7243745 52 - 22036055 ------UGUUCGCUGUUCACC-----UAGUGCUAAUCAUCACGAUUAGCCGCAUUGAUAAGGC ------.....(((..(((..-----..(.(((((((.....))))))))....)))...))) ( -13.10, z-score = -1.53, R) >droVir3.scaffold_12963 4820263 55 + 20206255 ---UGUAAGUAUUUGUUCACU-----UAGCGCUAAUCACUUCGAUUAGCCGCAUUGAUAUGGC ---..(((((........)))-----))(((((((((.....)))))).)))........... ( -12.70, z-score = -1.39, R) >droMoj3.scaffold_6500 13990195 55 + 32352404 -----UACAUGCAUUUAUAUAUA---UAUUUCUAAUCAGAAUGAUUAGCCGCAUUGAUAAGAC -----...((((...(((....)---))...((((((.....))))))..))))......... ( -6.00, z-score = -0.18, R) >droGri2.scaffold_15126 5037947 58 - 8399593 UGGAUUAAGUGCAUGUUCACU-----UAGCGCUAAUCACAUCGAUUAGCCGCAUUGAUAAGGC .....((((((......))))-----))(((((((((.....)))))).)))........... ( -15.20, z-score = -1.64, R) >consensus ______UGUUCCCUGUUCACC_____UAGUGCUAAUCAUCACGAUUAGCCGCAUUGAUAAGGC ............................(((((((((.....)))))).)))........... ( -8.22 = -8.30 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:44 2011