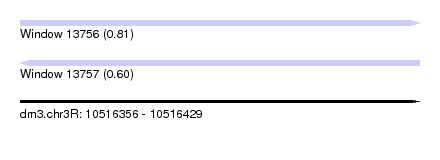
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,516,356 – 10,516,429 |
| Length | 73 |
| Max. P | 0.814217 |

| Location | 10,516,356 – 10,516,429 |
|---|---|
| Length | 73 |
| Sequences | 5 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Shannon entropy | 0.15858 |
| G+C content | 0.41370 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -17.24 |
| Energy contribution | -16.44 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
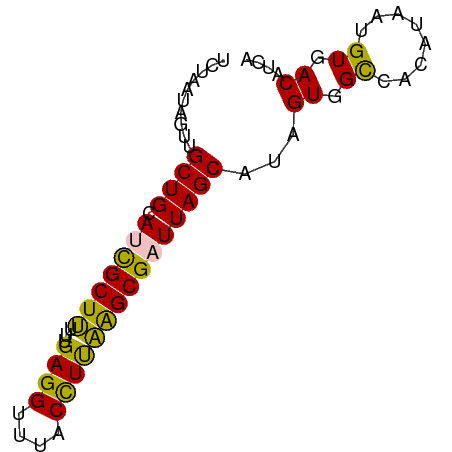
>dm3.chr3R 10516356 73 + 27905053 UCUAAUAGUUGCUGCAUCGCUCUUUGAGGUUUACUUUGAGCGAUUAGCAUAGUGGCCACAUAAUGUGACAUCA .........(((((.(((((((...(((.....))).))))))))))))..(((..(((.....))).))).. ( -21.70, z-score = -2.09, R) >droSim1.chr3R 16570596 73 - 27517382 UCUAAUAGUUGCUGCAACGCUUUUUGAGGUUUACCUUAAGCGAUUAGCAUAGUGGCCACAUAAUGUGACAUCA .........(((((...((((...(((((....)))))))))..)))))..(((..(((.....))).))).. ( -16.90, z-score = -0.82, R) >droSec1.super_0 9703822 73 - 21120651 UCUAAUAGUUGCUGCAACGCUUUUUGAGGUUUACCUUAAGCGAUUAGCAUAGUGGUCACAUAAUGUGACACCA .........(((((...((((...(((((....)))))))))..))))).....(((((.....))))).... ( -20.40, z-score = -2.24, R) >droYak2.chr3R 12915666 73 - 28832112 UCUAAUAGUGGCUGCAUCGCUUUUUGAGGUUUACCUCAGGCGAUUAGCAUAGUCGCCACAUAAUGUGACAUCA ..........((((.((((((...(((((....)))))))))))))))...(((((........))))).... ( -24.70, z-score = -3.34, R) >droEre2.scaffold_4770 10050844 73 + 17746568 UCUAAUAGUGGCUGCAUUGCUUGUUGAGGUUUACCUCAAGCGAUUAGCACUGUGGCAACAUAAUGUGACAUCA ......((((.(((.((((((...(((((....))))))))))))))))))((.(((......))).)).... ( -22.10, z-score = -1.65, R) >consensus UCUAAUAGUUGCUGCAUCGCUUUUUGAGGUUUACCUUAAGCGAUUAGCAUAGUGGCCACAUAAUGUGACAUCA ..........((((.(((((((...((((....)))))))))))))))...((.((........)).)).... (-17.24 = -16.44 + -0.80)

| Location | 10,516,356 – 10,516,429 |
|---|---|
| Length | 73 |
| Sequences | 5 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 90.96 |
| Shannon entropy | 0.15858 |
| G+C content | 0.41370 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.54 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10516356 73 - 27905053 UGAUGUCACAUUAUGUGGCCACUAUGCUAAUCGCUCAAAGUAAACCUCAAAGAGCGAUGCAGCAACUAUUAGA ....(((((.....))))).....((((.(((((((...............)))))))..))))......... ( -19.76, z-score = -2.43, R) >droSim1.chr3R 16570596 73 + 27517382 UGAUGUCACAUUAUGUGGCCACUAUGCUAAUCGCUUAAGGUAAACCUCAAAAAGCGUUGCAGCAACUAUUAGA ....(((((.....))))).....((((...(((((.(((....)))....)))))....))))......... ( -16.60, z-score = -1.07, R) >droSec1.super_0 9703822 73 + 21120651 UGGUGUCACAUUAUGUGACCACUAUGCUAAUCGCUUAAGGUAAACCUCAAAAAGCGUUGCAGCAACUAUUAGA (((((((((.....)))).)))))((((...(((((.(((....)))....)))))....))))......... ( -20.10, z-score = -2.51, R) >droYak2.chr3R 12915666 73 + 28832112 UGAUGUCACAUUAUGUGGCGACUAUGCUAAUCGCCUGAGGUAAACCUCAAAAAGCGAUGCAGCCACUAUUAGA ...((((((.....)))))).....(((.(((((.(((((....)))))....)))))..))).......... ( -22.20, z-score = -2.93, R) >droEre2.scaffold_4770 10050844 73 - 17746568 UGAUGUCACAUUAUGUUGCCACAGUGCUAAUCGCUUGAGGUAAACCUCAACAAGCAAUGCAGCCACUAUUAGA ..............(((((.....((((....(.((((((....))))))).))))..))))).......... ( -15.20, z-score = -0.18, R) >consensus UGAUGUCACAUUAUGUGGCCACUAUGCUAAUCGCUUAAGGUAAACCUCAAAAAGCGAUGCAGCAACUAUUAGA ....(((((.....)))))......(((.(((((((.(((....)))....)))))))..))).......... (-14.82 = -15.54 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:39 2011