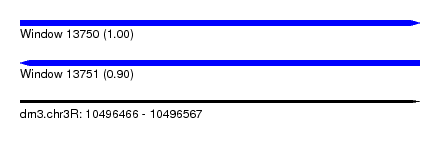
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,496,466 – 10,496,567 |
| Length | 101 |
| Max. P | 0.998275 |

| Location | 10,496,466 – 10,496,567 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 59.64 |
| Shannon entropy | 0.69377 |
| G+C content | 0.54275 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -18.60 |
| Energy contribution | -17.92 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
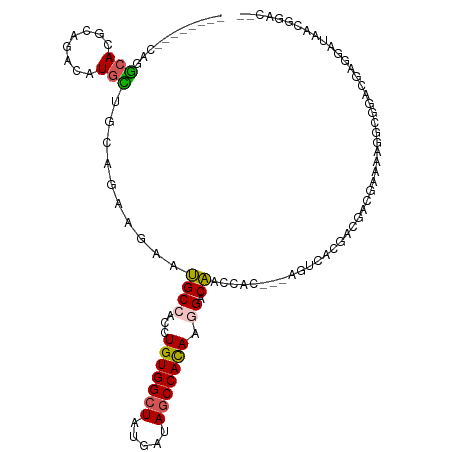
>dm3.chr3R 10496466 101 + 27905053 --GUCCGUUAUCCCCGUCAGCCUUUUCGUAGUCGUGACU---AUGGUCGUCCCUGUGGUUAUCAUAGCCACAGGUGGCGUUCGUCGGAUGCAUGUCUGCCUGCCUG-------- --(((((........(((((.((......)).).)))).---(((..((((((((((((((...)))))))))).))))..))))))))(((.(....).)))...-------- ( -33.50, z-score = -2.54, R) >droEre2.scaffold_4770 10030777 101 + 17746568 --GUCCGUUAUCCUCGUCAGCCUUUUCGUAGUCGUGGCU---GUGGUCGUCCCUGUGGCUAUCAUAGCCACAGGUGGCGUUCGUUGGAUGCAUGUCAGUCUGCCUG-------- --...((((((((....(((((....((....)).))))---).(..((((((((((((((...)))))))))).))))..)...))))).)))............-------- ( -37.40, z-score = -3.42, R) >droWil1.scaffold_181130 1283304 79 - 16660200 ------------------------UUCCCUGUCAUGGUU---GUGGUUGUCCUUGUGGCUGUCAUAGCCACAGGUGGCAUUCUUUUGCACCAUAUCUGCGUGACUG-------- ------------------------......((((((..(---(((((((((((((((((((...)))))))))).)))).........))))))....))))))..-------- ( -30.20, z-score = -3.19, R) >droMoj3.scaffold_6540 31210151 106 + 34148556 UUCCACCUCAUCCUCCUCCAGAUUCUCGCCGGGCCAAUUUUCAUAACUGCCUUUGUGGCUGUCGUAGCCACAGGUGGCAUUCUGCUGCAGCAUAUCGGCGUGCCUA-------- ........................(.((((((...............((((((((((((((...)))))))))).))))...(((....)))..)))))).)....-------- ( -30.00, z-score = -0.73, R) >droGri2.scaffold_14906 11928927 111 - 14172833 CUCCUGCUUAUGUUCCAUCCCGCUCUGCUCGUGUGGACU---GUGGUUGUCAGUAUGGCUAUCGUAGCCACAGGUGGCAUUCUGCUGCAAGAUGUCCGCGUGUCUGUUAUCCAA ........(((((..((((((((.(.....).)))).((---(((((((.(.(((....))).))))))))))(..((.....))..)..))))...)))))............ ( -28.30, z-score = 0.80, R) >consensus __CCCCCUUAUCCUCCUCAGCCUUUUCGUAGUCGUGACU___GUGGUUGUCCCUGUGGCUAUCAUAGCCACAGGUGGCAUUCUUCUGCAGCAUGUCUGCGUGCCUG________ ............................................((.((((((((((((((...)))))))))).)))).))................................ (-18.60 = -17.92 + -0.68)



| Location | 10,496,466 – 10,496,567 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 59.64 |
| Shannon entropy | 0.69377 |
| G+C content | 0.54275 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -10.32 |
| Energy contribution | -9.92 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10496466 101 - 27905053 --------CAGGCAGGCAGACAUGCAUCCGACGAACGCCACCUGUGGCUAUGAUAACCACAGGGACGACCAU---AGUCACGACUACGAAAAGGCUGACGGGGAUAACGGAC-- --------.......(((....)))((((..((...(((.(((((((.........)))))))(((......---.))).............)))...)).)))).......-- ( -27.80, z-score = -1.53, R) >droEre2.scaffold_4770 10030777 101 - 17746568 --------CAGGCAGACUGACAUGCAUCCAACGAACGCCACCUGUGGCUAUGAUAGCCACAGGGACGACCAC---AGCCACGACUACGAAAAGGCUGACGAGGAUAACGGAC-- --------...(((........)))((((......((.(.((((((((((...))))))))))).))....(---((((.............)))))....)))).......-- ( -33.32, z-score = -4.42, R) >droWil1.scaffold_181130 1283304 79 + 16660200 --------CAGUCACGCAGAUAUGGUGCAAAAGAAUGCCACCUGUGGCUAUGACAGCCACAAGGACAACCAC---AACCAUGACAGGGAA------------------------ --------..((((........(((((((......)))..(((((((((.....)))))).)))...)))).---.....))))......------------------------ ( -21.14, z-score = -1.25, R) >droMoj3.scaffold_6540 31210151 106 - 34148556 --------UAGGCACGCCGAUAUGCUGCAGCAGAAUGCCACCUGUGGCUACGACAGCCACAAAGGCAGUUAUGAAAAUUGGCCCGGCGAGAAUCUGGAGGAGGAUGAGGUGGAA --------......((((..(((.((.(..((((.((((...(((((((.....)))))))..(((....((....))..))).))))....))))..).)).))).))))... ( -34.30, z-score = -1.09, R) >droGri2.scaffold_14906 11928927 111 + 14172833 UUGGAUAACAGACACGCGGACAUCUUGCAGCAGAAUGCCACCUGUGGCUACGAUAGCCAUACUGACAACCAC---AGUCCACACGAGCAGAGCGGGAUGGAACAUAAGCAGGAG ...............((.(.((((((((.((.....))...(((((((((...)))))..((((.......)---)))........)))).))))))))...)....))..... ( -24.60, z-score = 0.15, R) >consensus ________CAGGCACGCAGACAUGCUGCAGAAGAAUGCCACCUGUGGCUAUGAUAGCCACAAGGACAACCAC___AGUCACGACGACGAAAAGGCGGACGAGGAUAACGGAC__ ...........(((........)))..........((((...(((((((.....))))))).)).))............................................... (-10.32 = -9.92 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:34 2011