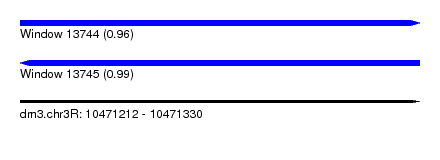
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,471,212 – 10,471,330 |
| Length | 118 |
| Max. P | 0.987293 |

| Location | 10,471,212 – 10,471,330 |
|---|---|
| Length | 118 |
| Sequences | 13 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 73.42 |
| Shannon entropy | 0.60905 |
| G+C content | 0.56819 |
| Mean single sequence MFE | -34.01 |
| Consensus MFE | -15.99 |
| Energy contribution | -16.42 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10471212 118 + 27905053 --CAUAAAAGUUUC-ACCGUUUUGAAACUUACCAGCAUCGUUAGUGCCUAUCAGCUCCGACUCGCUGAGACCGCCCAAUCCGGUAUCCUCGGAACUGGCGUAUGCCGAGCUGUUCUCGCAA --.....(((((((-(......))))))))....(((((((((((.((..(((((........))))).((((.......))))......)).))))))).))))((((.....))))... ( -35.00, z-score = -2.39, R) >droSim1.chr3R 16525703 111 - 27517382 ---------GUCCC-ACCGUUUUGAAACUUACCAGCAUCGUUAGCGCCUAUCAGCUCCGACUCGCUGAGACCGCCCAGUCCGGUAUCCUCGGAACUGGCGUAUGCCGAGCUGUUCUCGCAA ---------.....-.................((((.(((...(((....(((((........)))))...)))(((((((((.....)))).))))).......)))))))......... ( -29.10, z-score = -0.16, R) >droSec1.super_0 9554497 111 - 21120651 ---------GUCCC-ACCGUUUUGAAACUUACCAGCAUCGUUAGCGCCUAUCAGCUCCGACUCGCUGAGACCGCCCAGUCCGGUAUCCUCGGAACUGGCGUAUGCCGAGCUGUUCUCGCAA ---------.....-.................((((.(((...(((....(((((........)))))...)))(((((((((.....)))).))))).......)))))))......... ( -29.10, z-score = -0.16, R) >droYak2.chr3R 12869830 119 - 28832112 --CAUAUAAAUUCCCAUCGUUUCGAAACUUACCAGCAUCGUUAGCGCCUAUCAGCUCCGACUCGCUGAGACCGCCCAGUCCGGUGUCCUCGGAACUGGCGUACGCCGAGCUGUUCUCGCAA --..............................((((.(((...(((((....((.(((((..(((((.(((......))))))))...))))).)))))))....)))))))......... ( -32.00, z-score = -0.87, R) >droEre2.scaffold_4770 10004882 120 + 17746568 UAUACGGAAGUAAC-AUAAAUUGGAAACUUACCAGCAUCGUUAGCGCCUAUCAGCUCCGACUCGCUGAGACCGCCCAGUCCGGUGUCCUCGGAACUGGCGUAUGCCGAGCUGUUCUCGCAA ..(((....)))..-.......(....)....((((.(((.(((((((....((.(((((..(((((.(((......))))))))...))))).))))))).)).)))))))......... ( -36.80, z-score = -1.16, R) >droAna3.scaffold_13340 22562567 102 - 23697760 -------------------UGAGCCCACUCACCAGCAUCGUUGGCGCCAAUCAGCUCGGACUCGCUCAGCCCUCCCAGUCCAGUGUCCUCUGAGCUGGCGUACGCCGAGCUAUUCUCGCAG -------------------((((....))))...((......((((....((((((((((..((((..((.......))..))))...))))))))))....))))(((.....))))).. ( -32.50, z-score = -0.91, R) >dp4.chr2 19954507 110 - 30794189 -----------UGAUGCUCCAAGUGUACGUACCAGCAUCGUUAGCGCCAAUCAGCUCCGAUUCGCUCAGGCCACCCAGUCCCGUGUCCUCGGAACUGGCGUACGCCGAGCUGUUCUCGCAU -----------.((((((....(((....))).))))))....(((.....((((((.((.....)).(((.(((((((.(((......))).))))).))..)))))))))....))).. ( -37.70, z-score = -2.30, R) >droPer1.super_0 9634518 110 + 11822988 -----------UGAUGCUCCAAGUGUACGUACCAGCAUCGUUAGCGCCAAUCAGCUCCGACUCGCUCAGGCCACCCAGUCCCGUGUCCUCGGAACUGGCGUACGCCGAGCUGUUCUCGCAU -----------.((((((....(((....))).))))))....(((.....((((((.((.....)).(((.(((((((.(((......))).))))).))..)))))))))....))).. ( -37.60, z-score = -2.05, R) >droWil1.scaffold_181130 1261021 96 - 16660200 -------------------------AACUUACCAGCAUCGUUAGCGCCCAUCAGCUCCGAUUCACUUAGGCCACCCAGGCCCGUGUCUUCGGAACUGGCAUAGGCUGAACUAUUAUCGCAG -------------------------.......((((...(((((.((......))(((((..(((...((((.....)))).)))...))))).)))))....)))).............. ( -27.10, z-score = -1.54, R) >droVir3.scaffold_13047 13126563 118 + 19223366 ---UGAACUAAAUCCUCUAGCUGUCCACUUACCAGCAUCAUUUGCGCCUAUCAGCUCCGACUCACUGAGGCCACCCAGGCCCGUAUCCUCGGAGCUGGCAUAGGCCGAGCUGUUUUCGCAG ---..............(((((............(((.....)))(((((((((((((((.....((.((((.....)))))).....)))))))))..))))))..)))))......... ( -39.80, z-score = -2.60, R) >droMoj3.scaffold_6540 31187969 120 + 34148556 -CGAAUAUAAUGCGAAUAUGUUUUCCACUUACCAGCAUCAUUAGCGCCUAUCAGCUCCGACUCACUCAGACCUCCCAGGCCCGUAUCUUCGGAGCUGGCAUAGGCCGAACUGUUCUCGCAG -.........(((((..((((((...........((.......))(((((((((((((((......(.(.((.....)).).).....)))))))))..)))))).)))).))..))))). ( -33.40, z-score = -2.16, R) >droGri2.scaffold_14906 11899917 114 - 14172833 -------CAAGGCUCAUCAGCCAUUCACUUACCCGCAUCAUUCGCGCCUAUCAGCUCCGACUCACUCAGACCGCCCAGGCCCGUAUCCUCGGAGCUGGCAUAGGCCGAGCUGUUCUCGCAU -------...((((....))))............((..((((((.(((((((((((((((......(.(.((.....)).).).....)))))))))..)))))))))).)).....)).. ( -36.30, z-score = -2.35, R) >anoGam1.chr2R 28539199 99 + 62725911 ---------------------CGCUUACCUAUCA-CAUCCUGCGUCGCCAACAGCUCGGAUUCCGACAGCCCACCAACACCGGUGUCUUCCGAGCUGGCGUACGCGGAGCUGUUCUCACAA ---------------------............(-((.((((((((((((...(((((((....((((.((..........)))))).)))))))))))).)))))).).)))........ ( -35.70, z-score = -3.34, R) >consensus __________UUCC_ACCGUUUGGAAACUUACCAGCAUCGUUAGCGCCUAUCAGCUCCGACUCGCUCAGACCGCCCAGUCCCGUGUCCUCGGAACUGGCGUACGCCGAGCUGUUCUCGCAA ...........................................(((.....((((((.........................((.((....)))).(((....)))))))))....))).. (-15.99 = -16.42 + 0.42)


| Location | 10,471,212 – 10,471,330 |
|---|---|
| Length | 118 |
| Sequences | 13 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 73.42 |
| Shannon entropy | 0.60905 |
| G+C content | 0.56819 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -23.47 |
| Energy contribution | -23.03 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10471212 118 - 27905053 UUGCGAGAACAGCUCGGCAUACGCCAGUUCCGAGGAUACCGGAUUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCACUAACGAUGCUGGUAAGUUUCAAAACGGU-GAAACUUUUAUG-- .........(((((((.....((((...((((.(....)))))...))))((((((((........)))))..))).....))).))))..((((((((......)-))))))).....-- ( -39.30, z-score = -1.63, R) >droSim1.chr3R 16525703 111 + 27517382 UUGCGAGAACAGCUCGGCAUACGCCAGUUCCGAGGAUACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUUCAAAACGGU-GGGAC--------- .((((((.....))).)))..(((((((.(((.(....))))))).))))((((((.((..(.((((((....(((((....).))))....)))))).)..)).)-)))))--------- ( -40.30, z-score = -1.34, R) >droSec1.super_0 9554497 111 + 21120651 UUGCGAGAACAGCUCGGCAUACGCCAGUUCCGAGGAUACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUUCAAAACGGU-GGGAC--------- .((((((.....))).)))..(((((((.(((.(....))))))).))))((((((.((..(.((((((....(((((....).))))....)))))).)..)).)-)))))--------- ( -40.30, z-score = -1.34, R) >droYak2.chr3R 12869830 119 + 28832112 UUGCGAGAACAGCUCGGCGUACGCCAGUUCCGAGGACACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUUCGAAACGAUGGGAAUUUAUAUG-- ...((((.....)))).....(((((((.(((.(....))))))).)))).(((((.((..((((((((....(((((....).))))....))))))))..)).))))).........-- ( -41.90, z-score = -1.27, R) >droEre2.scaffold_4770 10004882 120 - 17746568 UUGCGAGAACAGCUCGGCAUACGCCAGUUCCGAGGACACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUUCCAAUUUAU-GUUACUUCCGUAUA .((((.((((((((((((...(((((((.(((.(....))))))).))))((((((((........)))))..)))))...))).))))((((((.....))))))-.....))))))).. ( -38.60, z-score = -0.33, R) >droAna3.scaffold_13340 22562567 102 + 23697760 CUGCGAGAAUAGCUCGGCGUACGCCAGCUCAGAGGACACUGGACUGGGAGGGCUGAGCGAGUCCGAGCUGAUUGGCGCCAACGAUGCUGGUGAGUGGGCUCA------------------- ...........(((((((.(.(.((((..(((......)))..))))).).)))))))((((((..(((.((..(((.......)))..)).))))))))).------------------- ( -44.40, z-score = -1.86, R) >dp4.chr2 19954507 110 + 30794189 AUGCGAGAACAGCUCGGCGUACGCCAGUUCCGAGGACACGGGACUGGGUGGCCUGAGCGAAUCGGAGCUGAUUGGCGCUAACGAUGCUGGUACGUACACUUGGAGCAUCA----------- ..(((..(((((((((((.(((.(((((((((......)))))))))))))))(((.....))))))))).))..)))....(((((((((......)))...)))))).----------- ( -46.30, z-score = -2.79, R) >droPer1.super_0 9634518 110 - 11822988 AUGCGAGAACAGCUCGGCGUACGCCAGUUCCGAGGACACGGGACUGGGUGGCCUGAGCGAGUCGGAGCUGAUUGGCGCUAACGAUGCUGGUACGUACACUUGGAGCAUCA----------- ..(((..(((((((((((.(((.(((((((((......)))))))))))))))(((.....))))))))).))..)))....(((((((((......)))...)))))).----------- ( -45.40, z-score = -2.02, R) >droWil1.scaffold_181130 1261021 96 + 16660200 CUGCGAUAAUAGUUCAGCCUAUGCCAGUUCCGAAGACACGGGCCUGGGUGGCCUAAGUGAAUCGGAGCUGAUGGGCGCUAACGAUGCUGGUAAGUU------------------------- ..(((....((((...((((((..(((((((((...((((((((.....)))))..)))..)))))))))))))))))))....))).........------------------------- ( -36.40, z-score = -2.07, R) >droVir3.scaffold_13047 13126563 118 - 19223366 CUGCGAAAACAGCUCGGCCUAUGCCAGCUCCGAGGAUACGGGCCUGGGUGGCCUCAGUGAGUCGGAGCUGAUAGGCGCAAAUGAUGCUGGUAAGUGGACAGCUAGAGGAUUUAGUUCA--- (..(.....(((((((((((((..(((((((((...((((((((.....)))))..)))..))))))))))))))).....))).))))....)..)..(((((((...)))))))..--- ( -44.90, z-score = -1.23, R) >droMoj3.scaffold_6540 31187969 120 - 34148556 CUGCGAGAACAGUUCGGCCUAUGCCAGCUCCGAAGAUACGGGCCUGGGAGGUCUGAGUGAGUCGGAGCUGAUAGGCGCUAAUGAUGCUGGUAAGUGGAAAACAUAUUCGCAUUAUAUUCG- ..(((((.....))).((((((..(((((((((..((.((((((.....)))))).))...)))))))))))))))))..(((((((..(((.((.....)).)))..))))))).....- ( -45.90, z-score = -2.99, R) >droGri2.scaffold_14906 11899917 114 + 14172833 AUGCGAGAACAGCUCGGCCUAUGCCAGCUCCGAGGAUACGGGCCUGGGCGGUCUGAGUGAGUCGGAGCUGAUAGGCGCGAAUGAUGCGGGUAAGUGAAUGGCUGAUGAGCCUUG------- ((.((....((..(((((((((..(((((((((..((.((((((.....)))))).))...))))))))))))))).))).))...)).))........((((....))))...------- ( -44.10, z-score = -1.30, R) >anoGam1.chr2R 28539199 99 - 62725911 UUGUGAGAACAGCUCCGCGUACGCCAGCUCGGAAGACACCGGUGUUGGUGGGCUGUCGGAAUCCGAGCUGUUGGCGACGCAGGAUG-UGAUAGGUAAGCG--------------------- .(((....)))(((((((((.((((((((((((.((((((.(......).)).))))....)))))))...))))))))).))).)-)............--------------------- ( -37.80, z-score = -1.22, R) >consensus UUGCGAGAACAGCUCGGCGUACGCCAGUUCCGAGGACACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUUACAAACGGU_GGAA__________ .........(((((((.....((((((((((((.......((.((....)).)).......))))))))....))))....))).))))................................ (-23.47 = -23.03 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:30 2011