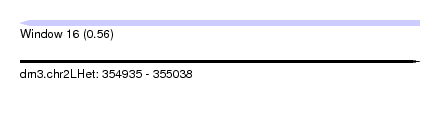
| Sequence ID | dm3.chr2LHet |
|---|---|
| Location | 354,935 – 355,038 |
| Length | 103 |
| Max. P | 0.560288 |

| Location | 354,935 – 355,038 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 51.37 |
| Shannon entropy | 0.68685 |
| G+C content | 0.36530 |
| Mean single sequence MFE | -16.04 |
| Consensus MFE | -8.24 |
| Energy contribution | -9.13 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.560288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
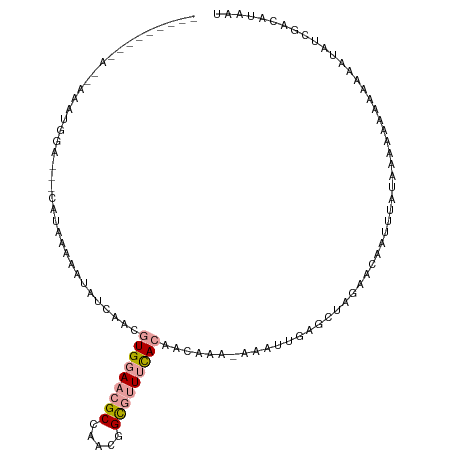
>dm3.chr2LHet 354935 103 - 368872 ---------AAAUAAUGGCUACCUUACGCAUAGCUACGUGGAACGCCAAUGGCGUUUCACAGCGCA-AACUUGAGCUAGCUCAAUUCCUACAUGAGAAGCAUAUCGACGUAAU ---------..............((((((...(((..((((((((((...)))))))))))))((.-...(((((....)))))((((.....).))))).....).))))). ( -28.70, z-score = -2.19, R) >anoGam1.chrU 44903631 112 - 59568033 AACUGAUGGAUCAUAUUCGAGAAAUAACUAUAUCAUACUGGAACGCAAACGGUGUUUUAAAAAAGA-AAAUUGAGCUAGAAAACUAUUUAAAGAAAAAAUAUGUUGACUUAAU ......(((.(((..(((((............))....((((((((.....)))))))).....))-)...))).)))...((((((((.......))))).)))........ ( -10.20, z-score = 0.81, R) >triCas2.chrUn_47 156243 97 + 241962 ------------AAAUGGA---CCUAAAAAUAUCAGCGUGGAAUGCCAACGGC-CUACACAACAAACUAAACGAGCUCCACCAAUUUACAUUACUCAACAAUAUCGACAUUAU ------------...((((---.((............(((....(((...)))-...))).............)).))))................................. ( -9.21, z-score = 0.11, R) >consensus _________A__AAAUGGA___CAUAAAAAUAUCAACGUGGAACGCCAACGGCGUUUCACAACAAA_AAAUUGAGCUAGAACAAUUUAUAAAAAAAAAAAAUAUCGACAUAAU .....................................(((((((((.....)))))))))..................................................... ( -8.24 = -9.13 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:04:52 2011