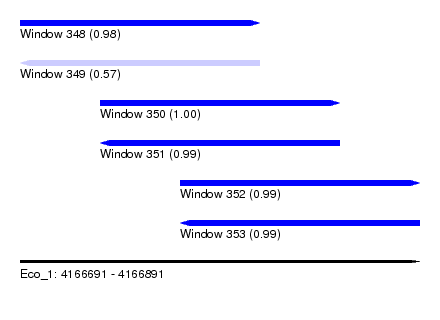
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,166,691 – 4,166,891 |
| Length | 200 |
| Max. P | 0.999515 |

| Location | 4,166,691 – 4,166,811 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -43.01 |
| Energy contribution | -40.82 |
| Covariance contribution | -2.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4166691 120 + 1 AUGCCCUGGCAGUCAGAGGCGAUGAAGGACGUGCUAAUCUGCGAUAAGCGUCGGUAAGGUGAUAUGAACCGUUAUAACCGGCGAUUUCCGAAUGGGGAAACCCAGUGUGUUUCGACACAC .(((((((.....))).)))).....(((..(((......))).....(((((((..(((.......)))......)))))))...)))...((((....))))((((((....)))))) ( -40.80) >Sfl_1 41 120 + 1 AUGCCCUGGCAGUCAGAGGCGAUGAAGGACGUGCUAAUCUGCGAUAAGCGUCGGUAAGGUGAUAUGAACCGUUAUAACCGGCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACAC ..((((((.....))).)))(((((((((..(((......))).....(((((((..(((.......)))......)))))))...)))...((((....))))......)))))).... ( -41.10) >Sty_1 41 120 + 1 AUGCCCUGGCAGUCAGAGGCGAUGAAGGACGUGCUAAUCUGCGAUAAGCGCCGGUAAGGUGAUAUGAACCGUUAUAACCGGCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACAC ..((((((.....))).)))(((((((((..(((......))).....(((((((..(((.......)))......)))))))...)))...((((....))))......)))))).... ( -43.80) >Ype_1 41 120 + 1 AUGCCUAGGCAGUCAGAGGCGAUGAAGGGCGUGCUAAUCUGCGAAAAGCGUCGGUAAGCUGAUAUGAAGCGUUAUAACCGACGAUACCCGAAUGGGGAAACCCAGUGCAAUACGUUGCAC .(((....))).......((((((..(((..(((......))......(((((((..(((.......)))......))))))))..)))(..((((....))))...)....)))))).. ( -40.70) >consensus AUGCCCUGGCAGUCAGAGGCGAUGAAGGACGUGCUAAUCUGCGAUAAGCGUCGGUAAGGUGAUAUGAACCGUUAUAACCGGCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACAC .(((((((.....))).)))).....(((..(((......))).....(((((((..(((.......)))......)))))))...)))...((((....))))((((((....)))))) (-43.01 = -40.82 + -2.19)


| Location | 4,166,691 – 4,166,811 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -34.25 |
| Energy contribution | -33.00 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4166691 120 - 1 GUGUGUCGAAACACACUGGGUUUCCCCAUUCGGAAAUCGCCGGUUAUAACGGUUCAUAUCACCUUACCGACGCUUAUCGCAGAUUAGCACGUCCUUCAUCGCCUCUGACUGCCAGGGCAU ((((((....))))))..(((((((......)))))))((..(((....((((............))))..((.....)).)))..))............(((.(((.....)))))).. ( -32.20) >Sfl_1 41 120 - 1 GUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGCCGGUUAUAACGGUUCAUAUCACCUUACCGACGCUUAUCGCAGAUUAGCACGUCCUUCAUCGCCUCUGACUGCCAGGGCAU ((((((....))))))..(((((((......)))))))((..(((....((((............))))..((.....)).)))..))............(((.(((.....)))))).. ( -32.30) >Sty_1 41 120 - 1 GUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGCCGGUUAUAACGGUUCAUAUCACCUUACCGGCGCUUAUCGCAGAUUAGCACGUCCUUCAUCGCCUCUGACUGCCAGGGCAU ((((((....))))))..(((((((......)))))))((((((......(((.......)))..))))))(((.(((...))).)))............(((.(((.....)))))).. ( -38.10) >Ype_1 41 120 - 1 GUGCAACGUAUUGCACUGGGUUUCCCCAUUCGGGUAUCGUCGGUUAUAACGCUUCAUAUCAGCUUACCGACGCUUUUCGCAGAUUAGCACGCCCUUCAUCGCCUCUGACUGCCUAGGCAU ((((((....))))))((((....))))...((((...((((((......(((.......)))..))))))(((..((...))..)))..))))......((((..(....)..)))).. ( -35.20) >consensus GUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGCCGGUUAUAACGGUUCAUAUCACCUUACCGACGCUUAUCGCAGAUUAGCACGUCCUUCAUCGCCUCUGACUGCCAGGGCAU ((((((....))))))..(((((((......)))))))((((((......(((.......)))..))))))((((...((((.((((..((........))...))))))))..)))).. (-34.25 = -33.00 + -1.25)



| Location | 4,166,731 – 4,166,851 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -45.74 |
| Energy contribution | -42.55 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.28 |
| Structure conservation index | 1.07 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4166731 120 + 1 GCGAUAAGCGUCGGUAAGGUGAUAUGAACCGUUAUAACCGGCGAUUUCCGAAUGGGGAAACCCAGUGUGUUUCGACACACUAUCAUUAACUGAAUCCAUAGGUUAAUGAGGCGAACCGGG ((.....))((((((..(((.......)))......)))))).....(((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))). ( -40.60) >Sfl_1 81 120 + 1 GCGAUAAGCGUCGGUAAGGUGAUAUGAACCGUUAUAACCGGCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACACUAUCAUUAACUGAAUCCAUAGGUUAAUGAGGCGAACCGGG ((.....))((((((..(((.......)))......)))))).....(((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))). ( -40.90) >Sty_1 81 120 + 1 GCGAUAAGCGCCGGUAAGGUGAUAUGAACCGUUAUAACCGGCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACACUAUCAUUAACUGAAUCCAUAGGUUAAUGAGGCGAACCGGG ((.....))((((((..(((.......)))......)))))).....(((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))). ( -43.60) >Ype_1 81 120 + 1 GCGAAAAGCGUCGGUAAGCUGAUAUGAAGCGUUAUAACCGACGAUACCCGAAUGGGGAAACCCAGUGCAAUACGUUGCACUAUCGUUAGAUGAAUACAUAGUCUAACGAGGCGAACCGGG ((.....))((((((..(((.......)))......))))))....((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......)))) ( -45.70) >consensus GCGAUAAGCGUCGGUAAGGUGAUAUGAACCGUUAUAACCGGCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACACUAUCAUUAACUGAAUCCAUAGGUUAAUGAGGCGAACCGGG ((.....))((((((..(((.......)))......))))))....((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......)))) (-45.74 = -42.55 + -3.19)


| Location | 4,166,731 – 4,166,851 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -35.06 |
| Energy contribution | -33.12 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.28 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4166731 120 - 1 CCCGGUUCGCCUCAUUAACCUAUGGAUUCAGUUAAUGAUAGUGUGUCGAAACACACUGGGUUUCCCCAUUCGGAAAUCGCCGGUUAUAACGGUUCAUAUCACCUUACCGACGCUUAUCGC .(((((.....((((((((...........))))))))((((((((....))))))))(((((((......))))))))))))......((((............))))........... ( -33.10) >Sfl_1 81 120 - 1 CCCGGUUCGCCUCAUUAACCUAUGGAUUCAGUUAAUGAUAGUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGCCGGUUAUAACGGUUCAUAUCACCUUACCGACGCUUAUCGC .(((((.....((((((((...........))))))))((((((((....))))))))(((((((......))))))))))))......((((............))))........... ( -33.20) >Sty_1 81 120 - 1 CCCGGUUCGCCUCAUUAACCUAUGGAUUCAGUUAAUGAUAGUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGCCGGUUAUAACGGUUCAUAUCACCUUACCGGCGCUUAUCGC ..((((..((.((((((((...........))))))))((((((((....))))))))(((((((......)))))))((((((......(((.......)))..))))))))..)))). ( -38.20) >Ype_1 81 120 - 1 CCCGGUUCGCCUCGUUAGACUAUGUAUUCAUCUAACGAUAGUGCAACGUAUUGCACUGGGUUUCCCCAUUCGGGUAUCGUCGGUUAUAACGCUUCAUAUCAGCUUACCGACGCUUUUCGC (((((......((((((((...((....)))))))))).(((((((....)))))))(((....)))..)))))...(((((((......(((.......)))..)))))))........ ( -36.30) >consensus CCCGGUUCGCCUCAUUAACCUAUGGAUUCAGUUAAUGAUAGUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGCCGGUUAUAACGGUUCAUAUCACCUUACCGACGCUUAUCGC ..((((..((.((((((((...........))))))))((((((((....))))))))(((((((......)))))))((((((......(((.......)))..))))))))..)))). (-35.06 = -33.12 + -1.94)


| Location | 4,166,771 – 4,166,891 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -44.08 |
| Energy contribution | -42.32 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.74 |
| Structure conservation index | 1.04 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4166771 120 + 1 GCGAUUUCCGAAUGGGGAAACCCAGUGUGUUUCGACACACUAUCAUUAACUGAAUCCAUAGGUUAAUGAGGCGAACCGGGGGAACUGAAACAUCUAAGUACCCCGAGGAAAAGAAAUCAA ..((((((((...(((....)))(((((((....))))))).(((((((((.........)))))))))..))..((.((((.(((..........))).))))..))....)))))).. ( -42.70) >Sfl_1 121 120 + 1 GCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACACUAUCAUUAACUGAAUCCAUAGGUUAAUGAGGCGAACCGGGGGAACUGAAACAUCUAAGUACCCCGAGGAAAAGAAAUCAA ..((((((((...(((....)))(((((((....))))))).(((((((((.........)))))))))..))..((.((((.(((..........))).))))..))....)))))).. ( -43.00) >Sty_1 121 120 + 1 GCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACACUAUCAUUAACUGAAUCCAUAGGUUAAUGAGGCGAACCGGGGGAACUGAAACAUCUAAGUACCCCGAGGAAAAGAAAUCAA ..((((((((...(((....)))(((((((....))))))).(((((((((.........)))))))))..))..((.((((.(((..........))).))))..))....)))))).. ( -43.00) >Ype_1 121 120 + 1 ACGAUACCCGAAUGGGGAAACCCAGUGCAAUACGUUGCACUAUCGUUAGAUGAAUACAUAGUCUAACGAGGCGAACCGGGGGAACUGAAACAUCUAAGUACCCCGAGGAAAAGAAAUCAA ..(((.(((....(((....)))(((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..)).......))).. ( -41.30) >consensus GCGAUUUCCGAAUGGGGAAACCCAGUGUGAUUCGUCACACUAUCAUUAACUGAAUCCAUAGGUUAAUGAGGCGAACCGGGGGAACUGAAACAUCUAAGUACCCCGAGGAAAAGAAAUCAA ..((((((((...(((....)))(((((((....))))))).(((((((((.........)))))))))..))..((.((((.(((..........))).))))..))....)))))).. (-44.08 = -42.32 + -1.75)


| Location | 4,166,771 – 4,166,891 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -36.69 |
| Energy contribution | -34.50 |
| Covariance contribution | -2.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 1.07 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4166771 120 - 1 UUGAUUUCUUUUCCUCGGGGUACUUAGAUGUUUCAGUUCCCCCGGUUCGCCUCAUUAACCUAUGGAUUCAGUUAAUGAUAGUGUGUCGAAACACACUGGGUUUCCCCAUUCGGAAAUCGC ..(((((((.......((((.(((..((....))))).)))).((...(((((((((((...........))))))))((((((((....)))))))))))..))......))))))).. ( -34.70) >Sfl_1 121 120 - 1 UUGAUUUCUUUUCCUCGGGGUACUUAGAUGUUUCAGUUCCCCCGGUUCGCCUCAUUAACCUAUGGAUUCAGUUAAUGAUAGUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGC ..(((((((.......((((.(((..((....))))).)))).((...(((((((((((...........))))))))((((((((....)))))))))))..))......))))))).. ( -34.80) >Sty_1 121 120 - 1 UUGAUUUCUUUUCCUCGGGGUACUUAGAUGUUUCAGUUCCCCCGGUUCGCCUCAUUAACCUAUGGAUUCAGUUAAUGAUAGUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGC ..(((((((.......((((.(((..((....))))).)))).((...(((((((((((...........))))))))((((((((....)))))))))))..))......))))))).. ( -34.80) >Ype_1 121 120 - 1 UUGAUUUCUUUUCCUCGGGGUACUUAGAUGUUUCAGUUCCCCCGGUUCGCCUCGUUAGACUAUGUAUUCAUCUAACGAUAGUGCAACGUAUUGCACUGGGUUUCCCCAUUCGGGUAUCGU ..............((((((.(((..((....)))))..))))))...(((((((((((...((....))))))))))((((((((....)))))))))))...(((....)))...... ( -32.70) >consensus UUGAUUUCUUUUCCUCGGGGUACUUAGAUGUUUCAGUUCCCCCGGUUCGCCUCAUUAACCUAUGGAUUCAGUUAAUGAUAGUGUGACGAAUCACACUGGGUUUCCCCAUUCGGAAAUCGC ..(((((((.......((((.(((..((....))))).)))).((...(((((((((((...........))))))))((((((((....)))))))))))..))......))))))).. (-36.69 = -34.50 + -2.19)

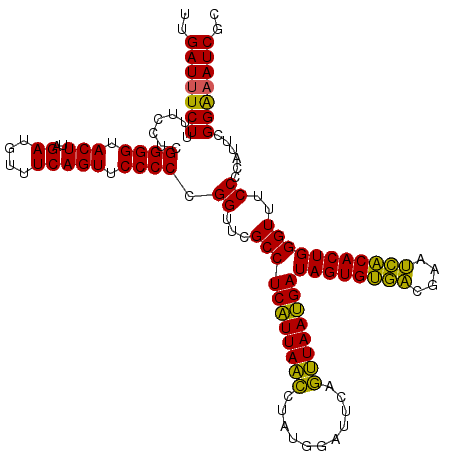
Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:07:06 2006