
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,164,536 – 4,165,136 |
| Length | 600 |
| Max. P | 0.994605 |

| Location | 4,164,536 – 4,164,656 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -30.13 |
| Energy contribution | -30.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164536 120 + 1 ACUGCUCUUUAACAAUUUAUCAGACAAUCUGUGUGGGCACUCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAG ....................(((.....))).(((..(((((..((((..((((((....((((....))))......)))).))..))))...)))))..)))((((....)))).... ( -30.90) >Sfl_1 1 120 + 1 ACUGCUCUUUAACAAUUUAUCAGACAAUCUGUGUGGGCACUCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAG ....................(((.....))).(((..(((((..((((..((((((....((((....))))......)))).))..))))...)))))..)))((((....)))).... ( -30.90) >Sty_1 1 120 + 1 ACUGCUCUUUAACAAUUUAUCAGACAAUCUGUGUGGGCACUCGAAGAUACGGAUUCUUAACGUCCUAGGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAG ....................(((.....))).(((..(((((..((((..((((((....((((....))))......)))).))..))))...)))))..)))((((....)))).... ( -28.60) >consensus ACUGCUCUUUAACAAUUUAUCAGACAAUCUGUGUGGGCACUCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAG ....................(((.....))).(((..(((((..((((..((((((....((((....))))......)))).))..))))...)))))..)))((((....)))).... (-30.13 = -30.13 + 0.00)



| Location | 4,164,536 – 4,164,656 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164536 120 - 1 CUUCGUAAUGAAUUACGUGUUCACUCUUGAGACUUGGUAUUCAUUUUUCGUCUUGCGACGUUAAGAAUCCGUAUCUUCGAGUGCCCACACAGAUUGUCUGAUAAAUUGUUAAAGAGCAGU (((..(((..(.(((.(((..(((((..((((..(((.((((.((...((((....))))..)))))))))..)))).)))))..))).(((.....))).))).)..)))..))).... ( -30.50) >Sfl_1 1 120 - 1 CUUCGUAAUGAAUUACGUGUUCACUCUUGAGACUUGGUAUUCAUUUUUCGUCUUGCGACGUUAAGAAUCCGUAUCUUCGAGUGCCCACACAGAUUGUCUGAUAAAUUGUUAAAGAGCAGU (((..(((..(.(((.(((..(((((..((((..(((.((((.((...((((....))))..)))))))))..)))).)))))..))).(((.....))).))).)..)))..))).... ( -30.50) >Sty_1 1 120 - 1 CUUCGUAAUGAAUUACGUGUUCACUCUUGAGACUUGGUAUUCAUUUUUCGUCCUAGGACGUUAAGAAUCCGUAUCUUCGAGUGCCCACACAGAUUGUCUGAUAAAUUGUUAAAGAGCAGU (((..(((..(.(((.(((..(((((..((((..(((.((((.((...((((....))))..)))))))))..)))).)))))..))).(((.....))).))).)..)))..))).... ( -32.00) >consensus CUUCGUAAUGAAUUACGUGUUCACUCUUGAGACUUGGUAUUCAUUUUUCGUCUUGCGACGUUAAGAAUCCGUAUCUUCGAGUGCCCACACAGAUUGUCUGAUAAAUUGUUAAAGAGCAGU (((..(((..(.(((.(((..(((((..((((..(((.((((.((...((((....))))..)))))))))..)))).)))))..))).(((.....))).))).)..)))..))).... (-31.00 = -31.00 + 0.00)

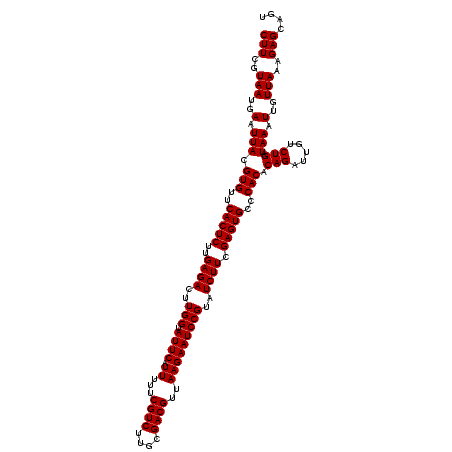
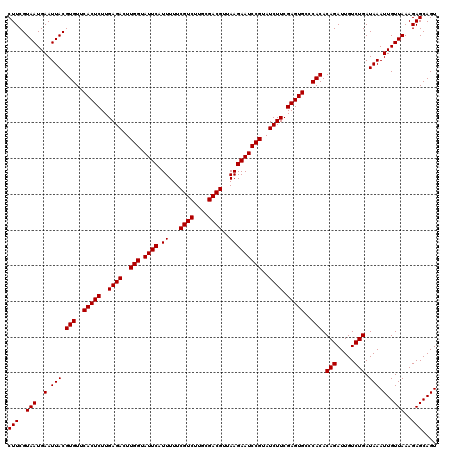
| Location | 4,164,576 – 4,164,696 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -29.35 |
| Energy contribution | -29.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.79 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.991003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164576 120 + 1 UCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCGUCAAACUUUUAAAUUGAAGAGUUU .....(((..((((((....((((....))))......)))).))..(.(((((((((((((.(((((....)))))..)))).)))).)))))))))((((((((......)))))))) ( -30.10) >Sfl_1 41 120 + 1 UCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUU .....(((..((((((....((((....))))......)))).))..(.(((((((((((((.(((((....)))))..)))).)))).)))))))))((((((((......)))))))) ( -29.80) >Sty_1 41 120 + 1 UCGAAGAUACGGAUUCUUAACGUCCUAGGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCAUUGAGCAUCAAACUUUUAAAUUGAAGAGUUU .....(((..((((((....((((....))))......)))).))..(.(((((((((((((.(((((....)))))..)))).)))).)))))))))((((((((......)))))))) ( -27.50) >consensus UCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUU .....(((..((((((....((((....))))......)))).))..(.(((((((((((((.(((((....)))))..)))).)))).)))))))))((((((((......)))))))) (-29.35 = -29.13 + -0.22)



| Location | 4,164,616 – 4,164,736 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -31.02 |
| Energy contribution | -30.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164616 120 + 1 AUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCGUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACAC ...(((.((.((((((((((((.(((((....)))))..)))).))))((((((((((((((((((......)))))))))))....)))))))))))))..))).((....))...... ( -31.80) >Sfl_1 81 120 + 1 AUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACAC ...(((.((.((((((((((((.(((((....)))))..)))).))))((((((((((((((((((......)))))))))))....)))))))))))))..))).((....))...... ( -31.50) >Sty_1 81 120 + 1 AUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCAUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACAC ...(((.((.((((((((((((.(((((....)))))..)))).)))).(((((((((((((((((......)))))))))))....)))))).))))))..))).((....))...... ( -31.10) >consensus AUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACAC ...(((.((.((((((((((((.(((((....)))))..)))).)))).(((((((((((((((((......)))))))))))....)))))).))))))..))).((....))...... (-31.02 = -30.80 + -0.22)

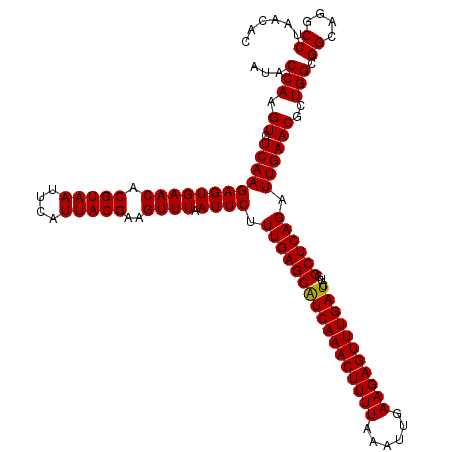
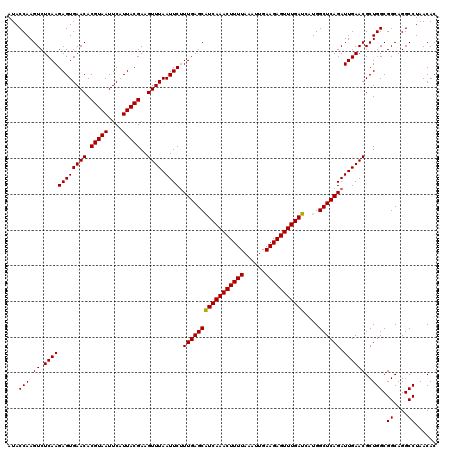
| Location | 4,164,656 – 4,164,776 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -37.04 |
| Energy contribution | -35.93 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164656 120 + 1 UUUAAUUCUUUGAGCGUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAACGGUAACAGGAAGAAGCUUGCUUCUUUG (((((((...((((((((((((((((......)))))))))))....)))))))))))).((((.((((..((.........))..))))..)))).....(((((((....))))))). ( -34.00) >Sfl_1 121 120 + 1 UUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAACGGUAACAGGAAGCAGCUUGCUGUUUCG (((((((...((((((((((((((((......)))))))))))....)))))))))))).((((.((((..((.........))..))))..)))).....(((((((....))))))). ( -35.70) >Sty_1 121 120 + 1 UUUAAUUCAUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAACGGUAACAGGAAGCAGCUUGCUGCUUCG .....(((((((((((((((((((((......)))))))))))....))))))..)))).((((.((((..((.........))..))))..)))).....(((((((....))))))). ( -38.30) >consensus UUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAACGGUAACAGGAAGCAGCUUGCUGCUUCG (((((((...((((((((((((((((......)))))))))))....)))))))))))).((((.((((..((.........))..))))..)))).....(((((((....))))))). (-37.04 = -35.93 + -1.10)


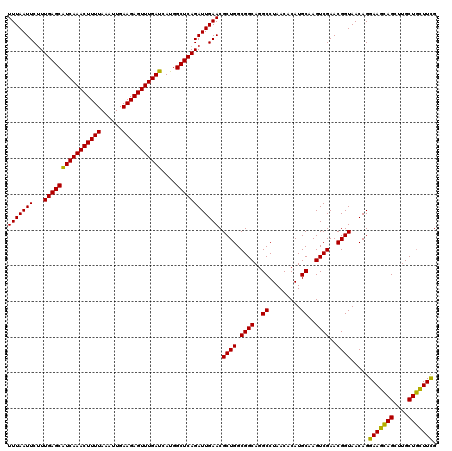
| Location | 4,164,736 – 4,164,856 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -37.80 |
| Energy contribution | -36.70 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.42 |
| Structure conservation index | 1.03 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164736 120 + 1 AUGCAAGUCGAACGGUAACAGGAAGAAGCUUGCUUCUUUGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUA .(((.(.(((..((.((.((((((((((....))))))).))).....)).))..))).)..))).(((((((....)).(((......)))))))).((((((....))))))...... ( -34.20) >Sfl_1 201 120 + 1 AUGCAAGUCGAACGGUAACAGGAAGCAGCUUGCUGUUUCGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUA .(((.(.(((..((.((.((((((((((....))))))).))).....)).))..))).)..))).(((((((....)).(((......)))))))).((((((....))))))...... ( -36.90) >Sty_1 201 120 + 1 AUGCAAGUCGAACGGUAACAGGAAGCAGCUUGCUGCUUCGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUGGCUAAUA .(((.(.(((..((.((.((((((((((....))))))).))).....)).))..))).)..))).(((((((....)).(((......)))))))).((((((....))))))...... ( -39.00) >consensus AUGCAAGUCGAACGGUAACAGGAAGCAGCUUGCUGCUUCGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUA .(((.(.(((..((.((.((((((((((....))))))).))).....)).))..))).)..))).(((((((....)).(((......)))))))).((((((....))))))...... (-37.80 = -36.70 + -1.10)



| Location | 4,164,776 – 4,164,896 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -41.22 |
| Energy contribution | -41.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164776 120 + 1 CUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGC .......((((((.....(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....))))))....((...((((....))))..)). ( -41.10) >Sfl_1 241 120 + 1 CUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGC .......((((((.....(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....))))))....((...((((....))))..)). ( -41.10) >Sty_1 241 120 + 1 CUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUGGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGC .......((((((.....(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....))))))....((...((((....))))..)). ( -40.80) >consensus CUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGC .......((((((.....(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....))))))....((...((((....))))..)). (-41.22 = -41.00 + -0.22)

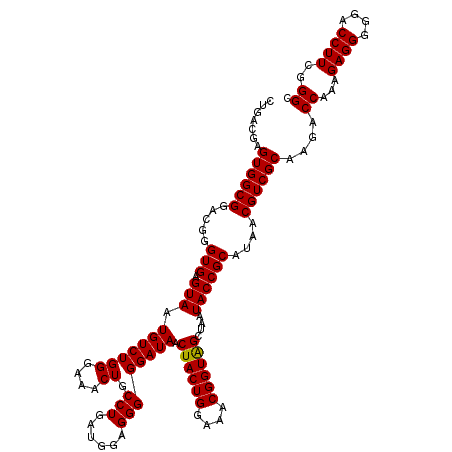
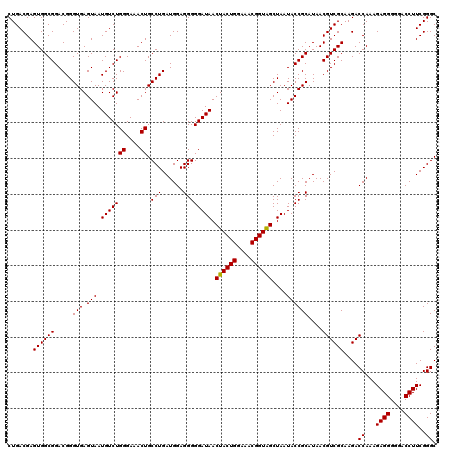
| Location | 4,164,816 – 4,164,936 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -48.43 |
| Consensus MFE | -48.87 |
| Energy contribution | -48.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.17 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164816 120 + 1 CCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAG .(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..((.(((....)))....))...... ( -48.30) >Sfl_1 281 120 + 1 CCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAG .(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..((.(((....)))....))...... ( -48.30) >Sty_1 281 120 + 1 CCUGAUGGAGGGGGAUAACUACUGGAAACGGUGGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCAGAUGUGCCCAGAUGGGAUUAGCUUGUUG .(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..((.(((....)))....))...... ( -48.70) >consensus CCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAG .(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..((.(((....)))....))...... (-48.87 = -48.43 + -0.44)



| Location | 4,164,856 – 4,164,976 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -52.23 |
| Consensus MFE | -50.93 |
| Energy contribution | -50.27 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164856 120 + 1 CCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGU ..(((((.((..((((((((...((((....))))..))..))))))...))..)))))((((..((((((.(((..((((((((.......)))))))))))...))))))...)))). ( -52.30) >Sfl_1 321 120 + 1 CCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGU ..(((((.((..((((((((...((((....))))..))..))))))...))..)))))((((..((((((.(((..((((((((.......)))))))))))...))))))...)))). ( -52.30) >Sty_1 321 120 + 1 CCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCAGAUGUGCCCAGAUGGGAUUAGCUUGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGU ..(((((.((((....))).....((((((.((....)))))))).......).)))))((((..((((((.(((..((((((((.......)))))))))))...))))))...)))). ( -52.10) >consensus CCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGU .(((((...((.((((((((...((((....))))..))..)))))).))...))))).((((..((((((.(((..((((((((.......)))))))))))...))))))...)))). (-50.93 = -50.27 + -0.66)

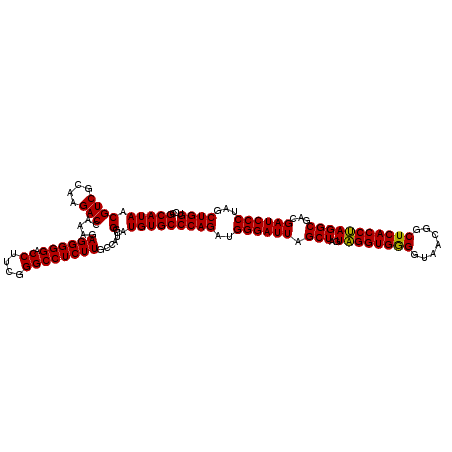

| Location | 4,164,936 – 4,165,056 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -47.53 |
| Consensus MFE | -44.72 |
| Energy contribution | -47.00 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164936 120 + 1 GUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAA (((.((((.((.((.....)).))....((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))))))).)))).))).. ( -47.50) >Sfl_1 401 120 + 1 GUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAA (((.((((.((.((.....)).))....((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))))))).)))).))).. ( -47.50) >Sty_1 401 120 + 1 GUGAGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAA (((((.......)))))....((((.(.((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))))))).).)))).... ( -47.60) >consensus GUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAA (((((.......)))))....((((.(.((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))))))).).)))).... (-44.72 = -47.00 + 2.28)


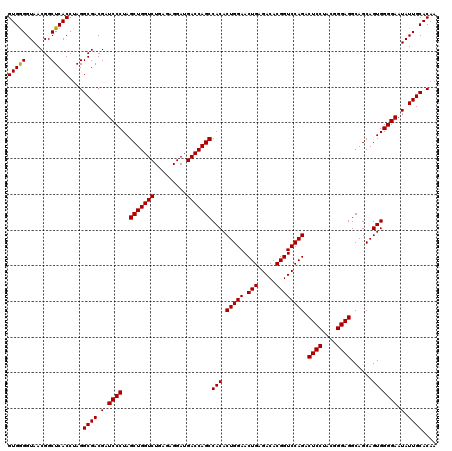
| Location | 4,165,016 – 4,165,136 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -45.80 |
| Consensus MFE | -41.70 |
| Energy contribution | -44.70 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4165016 120 + 1 CAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGCCGCGUGUAUGAAGAAGGCCUUCGGGUUGUAAAGUACUUUCAGCGGGGAGGAAGG ....((((....))))(((((((((.....)))))).....((((....))))..)))..((.(.((((..((((......((((....)))).....)))).....)))).).)).... ( -45.80) >Sfl_1 481 120 + 1 CAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGCCGCGUGUAUGAAGAAGGCCUUCGGGUUGUAAAGUACUUUCAGCGGGGAGGAAGG ....((((....))))(((((((((.....)))))).....((((....))))..)))..((.(.((((..((((......((((....)))).....)))).....)))).).)).... ( -45.80) >Sty_1 481 120 + 1 CAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGCCGCGUGUAUGAAGAAGGCCUUCGGGUUGUAAAGUACUUUCAGCGGGGAGGAAGG ....((((....))))(((((((((.....)))))).....((((....))))..)))..((.(.((((..((((......((((....)))).....)))).....)))).).)).... ( -45.80) >consensus CAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGCCGCGUGUAUGAAGAAGGCCUUCGGGUUGUAAAGUACUUUCAGCGGGGAGGAAGG ....((((....((..(((((((((.....)))))).....((((....))))..)))..))...((((..((((......((((....)))).....)))).....))))))))..... (-41.70 = -44.70 + 3.00)


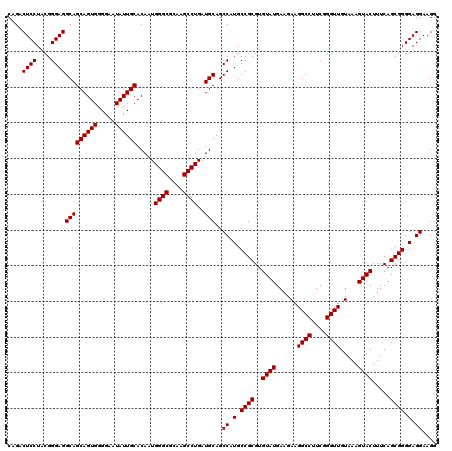
| Location | 4,164,760 – 4,164,880 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -39.63 |
| Consensus MFE | -36.71 |
| Energy contribution | -35.17 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164760 120 + 1 GAAGCUUGCUUCUUUGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAG ((((....))))(((((.((((....((((............(((((((....)).(((......)))))))).((((((....))))))......))))....)))))))))....... ( -39.20) >Hin_1 78 120 + 1 GAAGCUUGCUUUCUUGCUGACGAGUGGCGGACGGGUGAGUAAUGCUUGGGAAUCUGGCUUAUGGAGGGGGAUAACGACGGGAAACUGUCGCUAAUACCGCGUAUUAUCGGAAGAUGAAAG .((((((((((.(((((((........))).)))).)))))).))))((..((((..(((...)))..))))..((((((....))))))......)).....(((((....)))))... ( -36.40) >Ype_1 81 120 + 1 GUAGUUUACUACUUUGCCGGCGAGCGGCGGACGGGUGAGUAAUGUCUGGGGAUCUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUGACCUCGCAAGAGCAAAG ....((((((..(((((((.....)))))))..))))))....(((((.((((((.(((.....))).))))..((((((....))))))......)).)).)))(((....)))..... ( -43.30) >consensus GAAGCUUGCUUCUUUGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAUCUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACAUCGCAAGACCAAAG ...((((((((.(((((((.....))))))).)))))))).((((..((..((((.(((.....))).))))..((((((....))))))......))))))...(((....)))..... (-36.71 = -35.17 + -1.54)

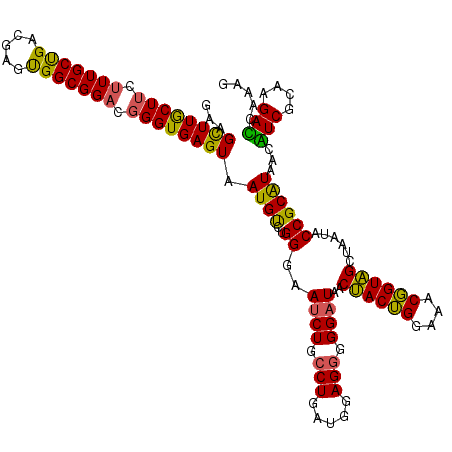
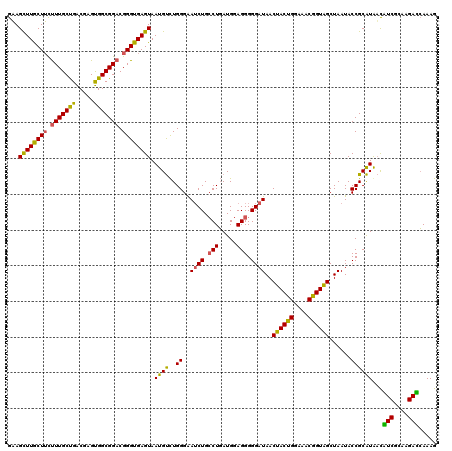
| Location | 4,164,800 – 4,164,920 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -51.33 |
| Consensus MFE | -52.01 |
| Energy contribution | -47.13 |
| Covariance contribution | -4.88 |
| Combinations/Pair | 1.32 |
| Mean z-score | -4.52 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164800 120 + 1 AAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGA ....((((((..((...(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..)).)))))) ( -53.20) >Hin_1 118 120 + 1 AAUGCUUGGGAAUCUGGCUUAUGGAGGGGGAUAACGACGGGAAACUGUCGCUAAUACCGCGUAUUAUCGGAAGAUGAAAGUGCGGGACUGAGAGGCCGCAUGCCAUAGGAUGAGCCCAAG ....((((((..((...(((((((.(.((..((.((((((....)))))).))...)).)...(((((....)))))..((((((..((....)))))))).)))))))..)).)))))) ( -49.70) >Ype_1 121 120 + 1 AAUGUCUGGGGAUCUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUGACCUCGCAAGAGCAAAGUGGGGGACCUUAGGGCCUCACGCCAUCGGAUGAACCCAGA ....((((((..((...(((((((..((((....((((((....))))))((((..((.(((...(((....)))....))).))....))))..))))...)))))))..)).)))))) ( -51.10) >consensus AAUGUCUGGGAAUCUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACAUCGCAAGACCAAAGUGGGGGACCUUAGGGCCUCAUGCCAUCGGAUGAGCCCAGA ....((((((..((...(((((((.(.((..((.((((((....)))))).))...)).).....(((....)))....((((((..((...)).)))))).)))))))..)).)))))) (-52.01 = -47.13 + -4.88)


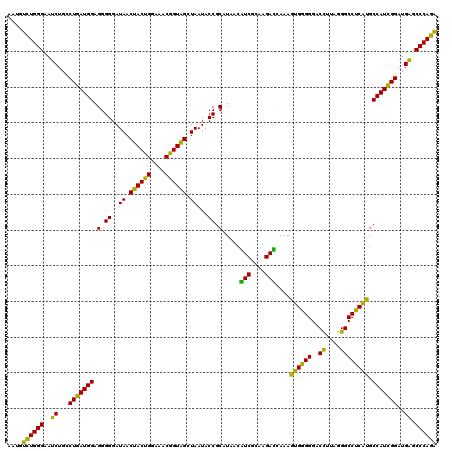
| Location | 4,164,800 – 4,164,920 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -31.01 |
| Energy contribution | -29.47 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164800 120 - 1 UCUGGGCACAUCCGAUGGCAAGAGGCCCGAAGGUCCCCCUCUUUGGUCUUGCGACGUUAUGCGGUAUUAGCUACCGUUUCCAGUAGUUAUCCCCCUCCAUCAGGCAGUUUCCCAGACAUU ((((((.((..((((((((((((...(((((((......))))))))))))).((.....((((((.....)))))).....)).............)))).))..))..)))))).... ( -37.20) >Hin_1 118 120 - 1 CUUGGGCUCAUCCUAUGGCAUGCGGCCUCUCAGUCCCGCACUUUCAUCUUCCGAUAAUACGCGGUAUUAGCGACAGUUUCCCGUCGUUAUCCCCCUCCAUAAGCCAGAUUCCCAAGCAUU ((((((...(((...((((.(((((..........)))))............(((((((((.((....(((....))).))))).))))))...........))))))).)))))).... ( -30.20) >Ype_1 121 120 - 1 UCUGGGUUCAUCCGAUGGCGUGAGGCCCUAAGGUCCCCCACUUUGCUCUUGCGAGGUCAUGCGGUAUUAGCUACCGUUUCCAGUAGUUAUCCCCCUCCAUCAGGCAGAUCCCCAGACAUU ((((((.((..((((((((((((((((....)))).....((((((....))))))))))))((...(((((((........))))))).)).....)))).))..))..)))))).... ( -41.90) >consensus UCUGGGCUCAUCCGAUGGCAUGAGGCCCCAAGGUCCCCCACUUUGAUCUUGCGACGUUAUGCGGUAUUAGCUACCGUUUCCAGUAGUUAUCCCCCUCCAUCAGGCAGAUUCCCAGACAUU ((((((.((..((((((((((((((((....))))..........(((....))).))))))((...(((((((........))))))).)).....)))).))..))..)))))).... (-31.01 = -29.47 + -1.55)



| Location | 4,164,840 – 4,164,960 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -40.95 |
| Energy contribution | -38.63 |
| Covariance contribution | -2.32 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164840 120 + 1 GAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCG .......((((((((.((((.....(((....))).....((((((.((....))))))))))((((((......)).)))))))))))))).((((((((.......)))))))).... ( -46.70) >Hin_1 158 120 + 1 GAAACUGUCGCUAAUACCGCGUAUUAUCGGAAGAUGAAAGUGCGGGACUGAGAGGCCGCAUGCCAUAGGAUGAGCCCAAGUGGGAUUAGGUAGUUGGUGGGGUAAAUGCCUACCAAGCCU ..(((((.(.(((((.((((...(((((....)))))..((((((..((....))))))))......((......))..)))).)))))))))))((((((.......))))))...... ( -39.80) >Ype_1 161 120 + 1 GAAACGGUAGCUAAUACCGCAUGACCUCGCAAGAGCAAAGUGGGGGACCUUAGGGCCUCACGCCAUCGGAUGAACCCAGAUGGGAUUAGCUAGUAGGUGGGGUAAUGGCUCACCUAGGCG .......((((((((.(((.(((..(((....)))....(((((((.((....)))))).))))))))).....(((....))))))))))).((((((((.......)))))))).... ( -44.80) >consensus GAAACGGUAGCUAAUACCGCAUAACAUCGCAAGACCAAAGUGGGGGACCUUAGGGCCUCAUGCCAUCGGAUGAGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAAAGGCUCACCUAGGCG .......((((((((.((((.....(((....)))....((((((..((...)).))))))))((((((......)).)))))))))))))).((((((((.......)))))))).... (-40.95 = -38.63 + -2.32)

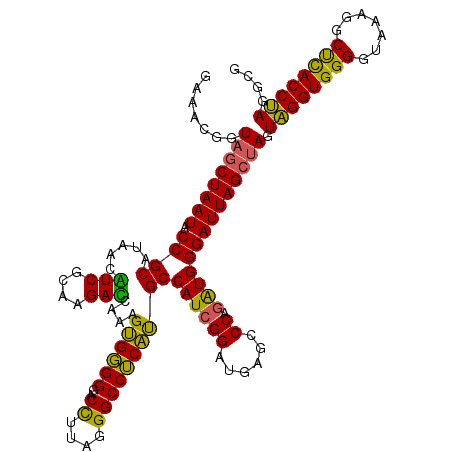
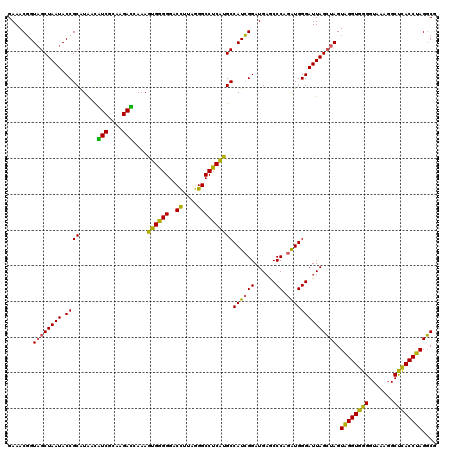
| Location | 4,164,880 – 4,165,000 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -47.37 |
| Consensus MFE | -43.14 |
| Energy contribution | -40.27 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164880 120 + 1 AGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUG ((((((.((....)))))))).....(((.((((.......((((((.(((..((((((((.......)))))))))))...))))))..(((((((.........)))))))))))))) ( -50.60) >Hin_1 198 120 + 1 UGCGGGACUGAGAGGCCGCAUGCCAUAGGAUGAGCCCAAGUGGGAUUAGGUAGUUGGUGGGGUAAAUGCCUACCAAGCCUGCGAUCUCUAGCUGGUCUGAGAGGAUGACCAGCCACACUG ..(((.....((((..(((((.((((.((......))..)))).)).((((..((((((((.......)))))))))))))))..)))).(((((((.........)))))))....))) ( -44.60) >Ype_1 201 120 + 1 UGGGGGACCUUAGGGCCUCACGCCAUCGGAUGAACCCAGAUGGGAUUAGCUAGUAGGUGGGGUAAUGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUG .((....)).(((((.....((((...((.(((.(((....))).))).))..((((((((.......)))))))))))).....)))))(((((((.........)))))))....... ( -46.90) >consensus UGGGGGACCUUAGGGCCUCAUGCCAUCGGAUGAGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAAAGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUG (((((..((...)).))))).......((......))....((((((.(((..((((((((.......)))))))))))...))))))..(((((((.........)))))))....... (-43.14 = -40.27 + -2.88)



| Location | 4,164,880 – 4,165,000 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -41.86 |
| Consensus MFE | -34.02 |
| Energy contribution | -34.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164880 120 - 1 CAGUGUGGCUGGUCAUCCUCUCAGACCAGCUAGGGAUCGUCGCCUAGGUGAGCCGUUACCCCACCUACUAGCUAAUCCCAUCUGGGCACAUCCGAUGGCAAGAGGCCCGAAGGUCCCCCU .....(((((((((.........)))))))))((((((.((((((((((((((.((..........))..))).....))))))))).........(((.....))).)).))))))... ( -43.00) >Hin_1 198 120 - 1 CAGUGUGGCUGGUCAUCCUCUCAGACCAGCUAGAGAUCGCAGGCUUGGUAGGCAUUUACCCCACCAACUACCUAAUCCCACUUGGGCUCAUCCUAUGGCAUGCGGCCUCUCAGUCCCGCA ..((((((((((((.........)))))))))(((..((((.(((.(((((................)))))....(((....)))..........))).))))..))).......))). ( -40.09) >Ype_1 201 120 - 1 CAGUGUGGCUGGUCAUCCUCUCAGACCAGCUAGGGAUCGUCGCCUAGGUGAGCCAUUACCCCACCUACUAGCUAAUCCCAUCUGGGUUCAUCCGAUGGCGUGAGGCCCUAAGGUCCCCCA .....(((((((((.........)))))))))((((((...(((((((((...........)))))........((.(((((.(((....)))))))).)).)))).....))))))... ( -42.50) >consensus CAGUGUGGCUGGUCAUCCUCUCAGACCAGCUAGGGAUCGUCGCCUAGGUGAGCCAUUACCCCACCUACUAGCUAAUCCCAUCUGGGCUCAUCCGAUGGCAUGAGGCCCCAAGGUCCCCCA .....(((((((((.........)))))))))((((((((((..((((((...........)))))).........(((....)))......))))(((.....)))....))))))... (-34.02 = -34.47 + 0.45)

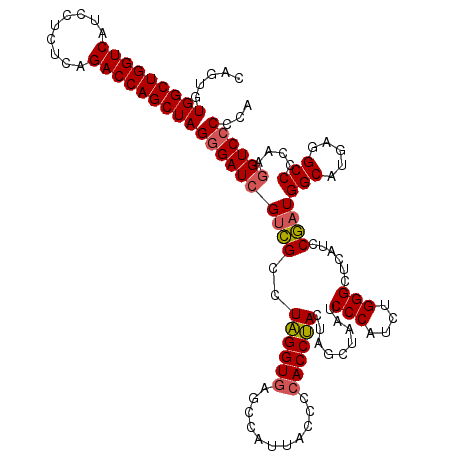

| Location | 4,164,920 – 4,165,040 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -50.80 |
| Consensus MFE | -49.28 |
| Energy contribution | -50.23 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164920 120 + 1 UGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGU .((((((.(((..((((((((.......)))))))))))...))))))..(((((((.........)))))))....(((((.(((.....)))))))).((((....))))........ ( -51.40) >Hin_1 238 120 + 1 UGGGAUUAGGUAGUUGGUGGGGUAAAUGCCUACCAAGCCUGCGAUCUCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGU .((((((((((..((((((((.......))))))))))))..))))))..(((((((.........)))))))....(((((.(((.....)))))))).((((....))))........ ( -49.60) >Ype_1 241 120 + 1 UGGGAUUAGCUAGUAGGUGGGGUAAUGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGU .((((((.(((..((((((((.......)))))))))))...))))))..(((((((.........)))))))....(((((.(((.....)))))))).((((....))))........ ( -51.40) >consensus UGGGAUUAGCUAGUAGGUGGGGUAAAGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGU .((((((.(((..((((((((.......)))))))))))...))))))..(((((((.........)))))))....(((((.(((.....)))))))).((((....))))........ (-49.28 = -50.23 + 0.95)

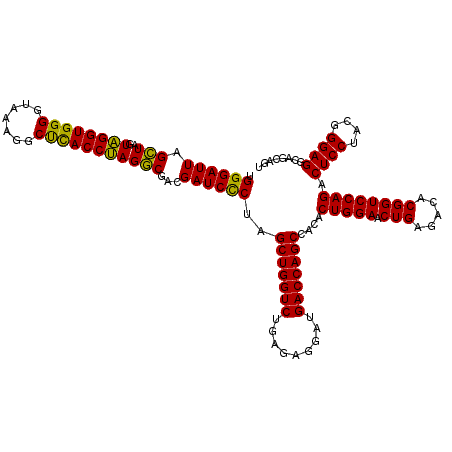

| Location | 4,164,960 – 4,165,080 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -50.43 |
| Consensus MFE | -48.67 |
| Energy contribution | -50.07 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164960 120 + 1 ACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAU ....((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......)))))))...(((((...(((((....))))).))))).... ( -51.20) >Hin_1 278 120 + 1 GCGAUCUCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCGCAAUGGGGGGAACCCUGACGCAGCCAU ((..((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......)))))))....((((...(((((....))))).))))))... ( -48.90) >Ype_1 281 120 + 1 ACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAU ....((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......)))))))...(((((...(((((....))))).))))).... ( -51.20) >consensus ACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAU ....((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......)))))))...(((((...(((((....))))).))))).... (-48.67 = -50.07 + 1.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:06:56 2006