
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,034,085 – 4,034,325 |
| Length | 240 |
| Max. P | 0.998023 |

| Location | 4,034,085 – 4,034,205 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -39.80 |
| Energy contribution | -37.03 |
| Covariance contribution | -2.76 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.12 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4034085 120 + 1 AAUACGGAGGGUGCAAGCGUUAAUCGGAAUUACUGGGCGUAAAGCGCACGCAGGCGGUUUGUUAAGUCAGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCUGAUACUGGCAAGC ...........(((..((((...((((.....))))(((.....)))))))..)))((((((((.((((((((((...((((.((......)).))))..))))))))))..)))))))) ( -40.90) >Hin_1 41 120 + 1 AAUACGGAGGGUGCGAGCGUUAAUCGGAAUAACUGGGCGUAAAGGGCACGCAGGCGGUUAUUUAAGUGAGGUGUGAAAGCCCCGGGCUUAACCUGGGAAUUGCAUUUCAGACUGGGUAAC ....(((.((((((((((.(((....(((((((((.((((.......))))...)))))))))...))).))((....))((((((.....))))))..))))))))....)))...... ( -34.70) >Sty_1 41 120 + 1 AAUACGGAGGGUGCAAGCGUUAAUCGGAAUUACUGGGCGUAAAGCGCACGCAGGCGGUCUGUCAAGUCGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUUCGAAACUGGCAGGC ...........(((..((((...((((.....))))(((.....)))))))..)))((((((((..(((((((((...((((.((......)).))))..)))))))))...)))))))) ( -43.40) >consensus AAUACGGAGGGUGCAAGCGUUAAUCGGAAUUACUGGGCGUAAAGCGCACGCAGGCGGUUUGUUAAGUCAGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUUUGAAACUGGCAAGC ...........(((..((((...((((.....))))(((.....)))))))..)))((((((((..((((((((......((((((.....))))))....))))))))...)))))))) (-39.80 = -37.03 + -2.76)


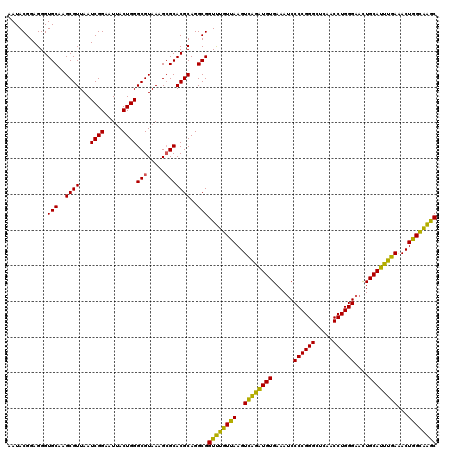
| Location | 4,034,125 – 4,034,245 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -40.16 |
| Energy contribution | -38.07 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4034125 120 + 1 AAAGCGCACGCAGGCGGUUUGUUAAGUCAGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCUGAUACUGGCAAGCUUGAGUCUCGUAGAGGGGGGUAGAAUUCCAGGUGUAGCGG ....((((((((((((((((((((.((((((((((...((((.((......)).))))..))))))))))..)))))))))...)))).......((((......))))..)))).))). ( -43.10) >Hin_1 81 120 + 1 AAAGGGCACGCAGGCGGUUAUUUAAGUGAGGUGUGAAAGCCCCGGGCUUAACCUGGGAAUUGCAUUUCAGACUGGGUAACUAGAGUACUUUAGGGAGGGGUAGAAUUCCACGUGUAGCGG ........((((.(((((((((((..(((((((..(....((((((.....))))))..)..)))))))...)))))))).............((((........)))).))).).))). ( -35.20) >Sty_1 81 120 + 1 AAAGCGCACGCAGGCGGUCUGUCAAGUCGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUUCGAAACUGGCAGGCUUGAGUCUUGUAGAGGGGGGUAGAAUUCCAGGUGUAGCGG ....((((((((((((((((((((..(((((((((...((((.((......)).))))..)))))))))...)))))))))...)))).......((((......))))..)))).))). ( -45.60) >consensus AAAGCGCACGCAGGCGGUUUGUUAAGUCAGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUUUGAAACUGGCAAGCUUGAGUCUUGUAGAGGGGGGUAGAAUUCCAGGUGUAGCGG ....((((((((((((((((((((..((((((((......((((((.....))))))....))))))))...)))))))))...))))........(((((...)))))..)))).))). (-40.16 = -38.07 + -2.10)

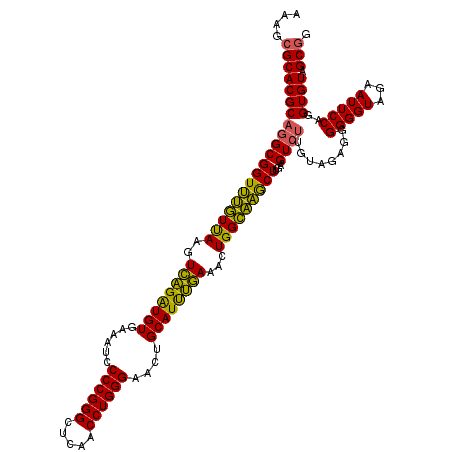

| Location | 4,034,205 – 4,034,325 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -47.03 |
| Consensus MFE | -47.61 |
| Energy contribution | -45.40 |
| Covariance contribution | -2.21 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.55 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4034205 120 + 1 UUGAGUCUCGUAGAGGGGGGUAGAAUUCCAGGUGUAGCGGUGAAAUGCGUAGAGAUCUGGAGGAAUACCGGUGGCGAAGGCGGCCCCCUGGACGAAGACUGACGCUCAGGUGCGAAAGCG ...((((((((..((((.((((...((((((((.(.(((......)))....).))))))))...))))(((.((....)).)))))))..))).)))))...........((....)). ( -51.30) >Hin_1 161 120 + 1 UAGAGUACUUUAGGGAGGGGUAGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAAUACCGAAGGCGAAGGCAGCCCCUUGGGAAUGUACUGACGCUCAUGUGCGAAAGCG ...((((((((((((.((((((...((((((((.(.(((......)))....).))))))))...))))....((....))..)))))))))...)))))...........((....)). ( -40.30) >Sty_1 161 120 + 1 UUGAGUCUUGUAGAGGGGGGUAGAAUUCCAGGUGUAGCGGUGAAAUGCGUAGAGAUCUGGAGGAAUACCGGUGGCGAAGGCGGCCCCCUGGACAAAGACUGACGCUCAGGUGCGAAAGCG ...((((((((..((((.((((...((((((((.(.(((......)))....).))))))))...))))(((.((....)).)))))))..)).))))))...........((....)). ( -49.50) >consensus UUGAGUCUUGUAGAGGGGGGUAGAAUUCCAGGUGUAGCGGUGAAAUGCGUAGAGAUCUGGAGGAAUACCGGUGGCGAAGGCGGCCCCCUGGACAAAGACUGACGCUCAGGUGCGAAAGCG ...((((((((...((((((((...((((((((.(.(((......)))....).))))))))...))))(((.((....)).)))))))..))).)))))...........((....)). (-47.61 = -45.40 + -2.21)

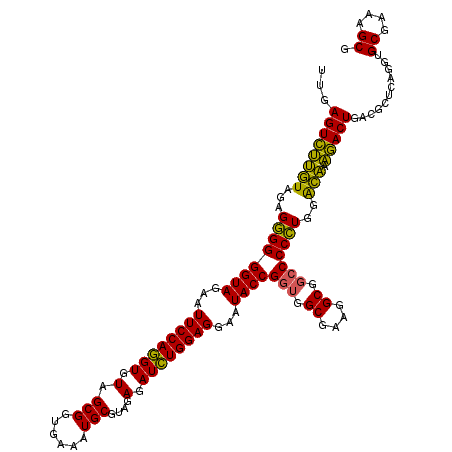
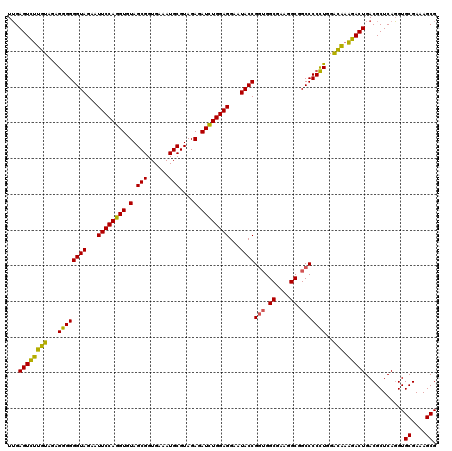
| Location | 4,034,205 – 4,034,325 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -29.32 |
| Energy contribution | -27.89 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.75 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4034205 120 - 1 CGCUUUCGCACCUGAGCGUCAGUCUUCGUCCAGGGGGCCGCCUUCGCCACCGGUAUUCCUCCAGAUCUCUACGCAUUUCACCGCUACACCUGGAAUUCUACCCCCCUCUACGAGACUCAA (((((........)))))..(((((.(((..(((((((.......)))...((((....(((((........((........)).....)))))....)))).))))..))))))))... ( -32.62) >Hin_1 161 120 - 1 CGCUUUCGCACAUGAGCGUCAGUACAUUCCCAAGGGGCUGCCUUCGCCUUCGGUAUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCUACCCCUCCCUAAAGUACUCUA (((((........)))))..(((((.......(((((.((((.........))))....(((((........((........)).....)))))......))))).......)))))... ( -24.86) >Sty_1 161 120 - 1 CGCUUUCGCACCUGAGCGUCAGUCUUUGUCCAGGGGGCCGCCUUCGCCACCGGUAUUCCUCCAGAUCUCUACGCAUUUCACCGCUACACCUGGAAUUCUACCCCCCUCUACAAGACUCAA (((((........)))))..((((((.((..(((((((.......)))...((((....(((((........((........)).....)))))....)))).))))..))))))))... ( -30.82) >consensus CGCUUUCGCACCUGAGCGUCAGUCUUUGUCCAGGGGGCCGCCUUCGCCACCGGUAUUCCUCCAGAUCUCUACGCAUUUCACCGCUACACCUGGAAUUCUACCCCCCUCUACAAGACUCAA (((((........)))))..(((((.(((..(((((((.......)))...((((....(((((........((........)).....)))))....)))).))))..))))))))... (-29.32 = -27.89 + -1.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:06:25 2006