
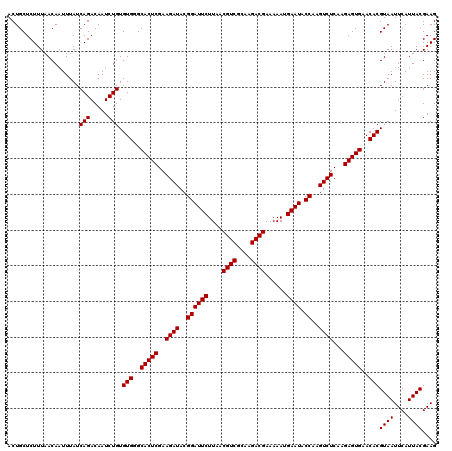
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,033,408 – 4,034,008 |
| Length | 600 |
| Max. P | 0.993102 |

| Location | 4,033,408 – 4,033,528 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -30.13 |
| Energy contribution | -30.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033408 120 + 1 ACUGCUCUUUAACAAUUUAUCAGACAAUCUGUGUGGGCACUCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAG ....................(((.....))).(((..(((((..((((..((((((....((((....))))......)))).))..))))...)))))..)))((((....)))).... ( -30.90) >Sfl_1 1 120 + 1 ACUGCUCUUUAACAAUUUAUCAGACAAUCUGUGUGGGCACUCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAG ....................(((.....))).(((..(((((..((((..((((((....((((....))))......)))).))..))))...)))))..)))((((....)))).... ( -30.90) >Sty_1 1 120 + 1 ACUGCUCUUUAACAAUUUAUCAGACAAUCUGUGUGGGCACUCGAAGAUACGGAUUCUUAACGUCCUAGGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAG ....................(((.....))).(((..(((((..((((..((((((....((((....))))......)))).))..))))...)))))..)))((((....)))).... ( -28.60) >consensus ACUGCUCUUUAACAAUUUAUCAGACAAUCUGUGUGGGCACUCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAG ....................(((.....))).(((..(((((..((((..((((((....((((....))))......)))).))..))))...)))))..)))((((....)))).... (-30.13 = -30.13 + 0.00)


| Location | 4,033,408 – 4,033,528 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033408 120 - 1 CUUCGUAAUGAAUUACGUGUUCACUCUUGAGACUUGGUAUUCAUUUUUCGUCUUGCGACGUUAAGAAUCCGUAUCUUCGAGUGCCCACACAGAUUGUCUGAUAAAUUGUUAAAGAGCAGU (((..(((..(.(((.(((..(((((..((((..(((.((((.((...((((....))))..)))))))))..)))).)))))..))).(((.....))).))).)..)))..))).... ( -30.50) >Sfl_1 1 120 - 1 CUUCGUAAUGAAUUACGUGUUCACUCUUGAGACUUGGUAUUCAUUUUUCGUCUUGCGACGUUAAGAAUCCGUAUCUUCGAGUGCCCACACAGAUUGUCUGAUAAAUUGUUAAAGAGCAGU (((..(((..(.(((.(((..(((((..((((..(((.((((.((...((((....))))..)))))))))..)))).)))))..))).(((.....))).))).)..)))..))).... ( -30.50) >Sty_1 1 120 - 1 CUUCGUAAUGAAUUACGUGUUCACUCUUGAGACUUGGUAUUCAUUUUUCGUCCUAGGACGUUAAGAAUCCGUAUCUUCGAGUGCCCACACAGAUUGUCUGAUAAAUUGUUAAAGAGCAGU (((..(((..(.(((.(((..(((((..((((..(((.((((.((...((((....))))..)))))))))..)))).)))))..))).(((.....))).))).)..)))..))).... ( -32.00) >consensus CUUCGUAAUGAAUUACGUGUUCACUCUUGAGACUUGGUAUUCAUUUUUCGUCUUGCGACGUUAAGAAUCCGUAUCUUCGAGUGCCCACACAGAUUGUCUGAUAAAUUGUUAAAGAGCAGU (((..(((..(.(((.(((..(((((..((((..(((.((((.((...((((....))))..)))))))))..)))).)))))..))).(((.....))).))).)..)))..))).... (-31.00 = -31.00 + 0.00)


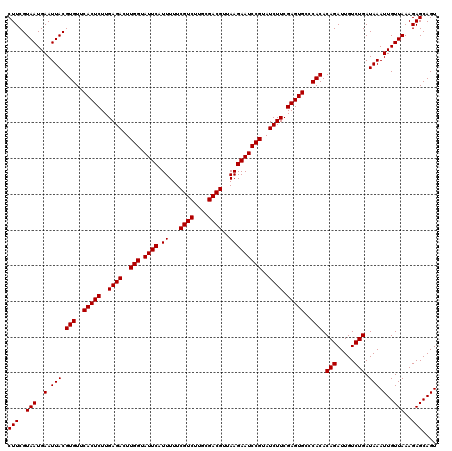
| Location | 4,033,448 – 4,033,568 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -29.35 |
| Energy contribution | -29.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.79 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.991003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033448 120 + 1 UCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCGUCAAACUUUUAAAUUGAAGAGUUU .....(((..((((((....((((....))))......)))).))..(.(((((((((((((.(((((....)))))..)))).)))).)))))))))((((((((......)))))))) ( -30.10) >Sfl_1 41 120 + 1 UCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUU .....(((..((((((....((((....))))......)))).))..(.(((((((((((((.(((((....)))))..)))).)))).)))))))))((((((((......)))))))) ( -29.80) >Sty_1 41 120 + 1 UCGAAGAUACGGAUUCUUAACGUCCUAGGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCAUUGAGCAUCAAACUUUUAAAUUGAAGAGUUU .....(((..((((((....((((....))))......)))).))..(.(((((((((((((.(((((....)))))..)))).)))).)))))))))((((((((......)))))))) ( -27.50) >consensus UCGAAGAUACGGAUUCUUAACGUCGCAAGACGAAAAAUGAAUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUU .....(((..((((((....((((....))))......)))).))..(.(((((((((((((.(((((....)))))..)))).)))).)))))))))((((((((......)))))))) (-29.35 = -29.13 + -0.22)

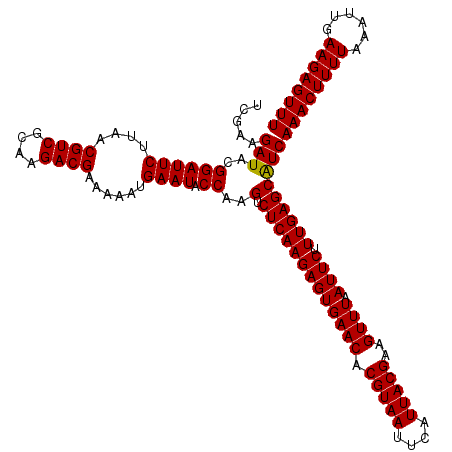
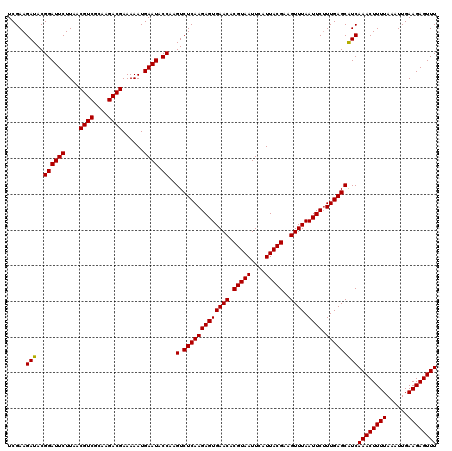
| Location | 4,033,488 – 4,033,608 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -31.02 |
| Energy contribution | -30.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033488 120 + 1 AUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCGUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACAC ...(((.((.((((((((((((.(((((....)))))..)))).))))((((((((((((((((((......)))))))))))....)))))))))))))..))).((....))...... ( -31.80) >Sfl_1 81 120 + 1 AUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACAC ...(((.((.((((((((((((.(((((....)))))..)))).))))((((((((((((((((((......)))))))))))....)))))))))))))..))).((....))...... ( -31.50) >Sty_1 81 120 + 1 AUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCAUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACAC ...(((.((.((((((((((((.(((((....)))))..)))).)))).(((((((((((((((((......)))))))))))....)))))).))))))..))).((....))...... ( -31.10) >consensus AUACCAAGUCUCAAGAGUGAACACGUAAUUCAUUACGAAGUUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACAC ...(((.((.((((((((((((.(((((....)))))..)))).)))).(((((((((((((((((......)))))))))))....)))))).))))))..))).((....))...... (-31.02 = -30.80 + -0.22)


| Location | 4,033,528 – 4,033,648 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -35.43 |
| Consensus MFE | -35.30 |
| Energy contribution | -34.53 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033528 120 + 1 UUUAAUUCUUUGAGCGUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAACGGUAACAGGAAGAAGCUUGCUUCUUUG (((((((...((((((((((((((((......)))))))))))....)))))))))))).((((.((((..((.........))..))))..)))).....(((((((....))))))). ( -34.00) >Sfl_1 121 119 + 1 UUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAACGGUAACAGGAAGC-GCUUGCUGUUUCG (((((((...((((((((((((((((......)))))))))))....))))))))))))((..((((((((((((..(...(((..........)))...)..)).-)))))))))).)) ( -34.00) >Sty_1 121 120 + 1 UUUAAUUCAUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAACGGUAACAGGAAGCAGCUUGCUGCUUCG .....(((((((((((((((((((((......)))))))))))....))))))..)))).((((.((((..((.........))..))))..)))).....(((((((....))))))). ( -38.30) >consensus UUUAAUUCUUUGAGCAUCAAACUUUUAAAUUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAACGGUAACAGGAAGCAGCUUGCUGCUUCG (((((((...((((((((((((((((......)))))))))))....)))))))))))).((((.((((..((.........))..))))..)))).....(((((((....))))))). (-35.30 = -34.53 + -0.77)



| Location | 4,033,648 – 4,033,768 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -40.22 |
| Energy contribution | -40.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033648 120 + 1 CUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGUACCUUCGGGC .......((((((.....(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....))))))....((...(((((...))))).)). ( -38.70) >Sfl_1 240 120 + 1 CUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGC .......((((((.....(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....))))))....((...((((....))))..)). ( -41.10) >Sty_1 241 120 + 1 CUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUGGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGC .......((((((.....(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....))))))....((...((((....))))..)). ( -40.80) >consensus CUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGC .......((((((.....(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....))))))....((...((((....))))..)). (-40.22 = -40.00 + -0.22)

| Location | 4,033,688 – 4,033,808 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -47.70 |
| Consensus MFE | -47.87 |
| Energy contribution | -47.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.01 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
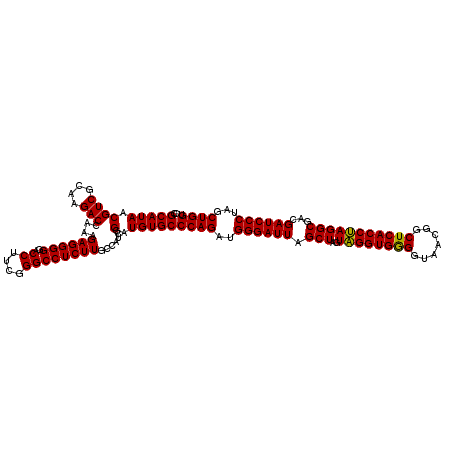
>Eco_1 4033688 120 + 1 CCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGUACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAG .(((((((((((((....((((((....))))))....((((.(.....(((....)))......).))))((....)))))))).)))))))..((.(((....)))....))...... ( -46.10) >Sfl_1 280 120 + 1 CCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAG .(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..((.(((....)))....))...... ( -48.30) >Sty_1 281 120 + 1 CCUGAUGGAGGGGGAUAACUACUGGAAACGGUGGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCAGAUGUGCCCAGAUGGGAUUAGCUUGUUG .(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..((.(((....)))....))...... ( -48.70) >consensus CCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAG .(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..((.(((....)))....))...... (-47.87 = -47.43 + -0.44)

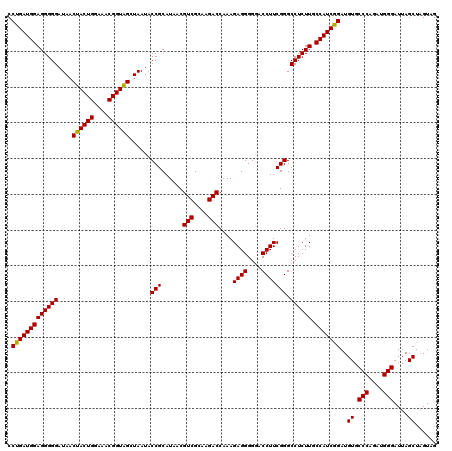
| Location | 4,033,728 – 4,033,848 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -51.43 |
| Consensus MFE | -49.93 |
| Energy contribution | -49.27 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033728 120 + 1 CCGCAUAACGUCGCAAGACCAAAGAGGGGUACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGU ..(((((.((..((((((((...(((((...))))).))..))))))...))..)))))((((..((((((.(((..((((((((.......)))))))))))...))))))...)))). ( -49.90) >Sfl_1 320 120 + 1 CCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGU ..(((((.((..((((((((...((((....))))..))..))))))...))..)))))((((..((((((.(((..((((((((.......)))))))))))...))))))...)))). ( -52.30) >Sty_1 321 120 + 1 CCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCAGAUGUGCCCAGAUGGGAUUAGCUUGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGU ..(((((.((((....))).....((((((.((....)))))))).......).)))))((((..((((((.(((..((((((((.......)))))))))))...))))))...)))). ( -52.10) >consensus CCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUAGGUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGU .(((((...((.((((((((...((((....))))..))..)))))).))...))))).((((..((((((.(((..((((((((.......)))))))))))...))))))...)))). (-49.93 = -49.27 + -0.66)

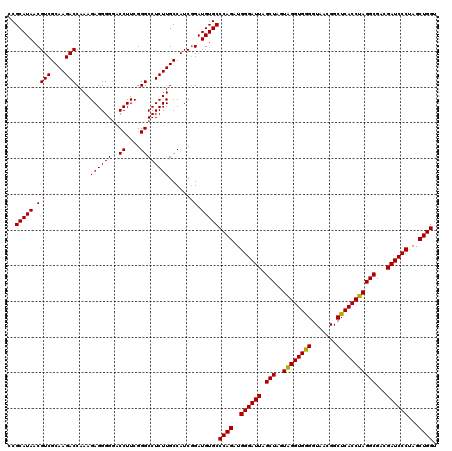
| Location | 4,033,808 – 4,033,928 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -47.53 |
| Consensus MFE | -44.72 |
| Energy contribution | -47.00 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033808 120 + 1 GUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAA (((.((((.((.((.....)).))....((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))))))).)))).))).. ( -47.50) >Sfl_1 400 120 + 1 GUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAA (((.((((.((.((.....)).))....((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))))))).)))).))).. ( -47.50) >Sty_1 401 120 + 1 GUGAGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAA (((((.......)))))....((((.(.((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))))))).).)))).... ( -47.60) >consensus GUGGGGUAACGGCUCACCUAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAA (((((.......)))))....((((.(.((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))))))).).)))).... (-44.72 = -47.00 + 2.28)

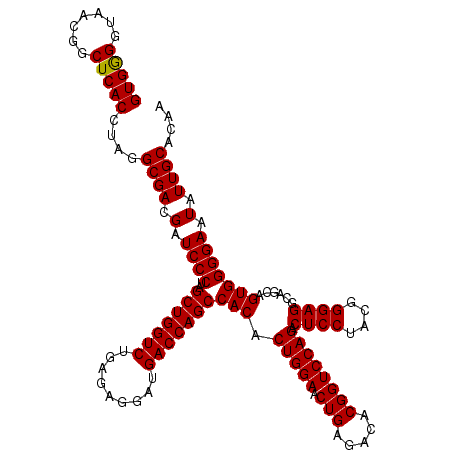
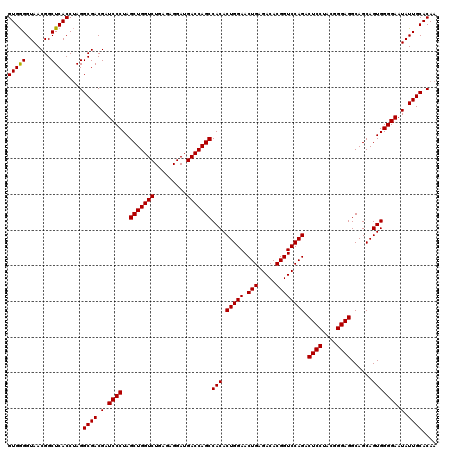
| Location | 4,033,888 – 4,034,008 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -45.80 |
| Consensus MFE | -41.70 |
| Energy contribution | -44.70 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033888 120 + 1 CAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGCCGCGUGUAUGAAGAAGGCCUUCGGGUUGUAAAGUACUUUCAGCGGGGAGGAAGG ....((((....))))(((((((((.....)))))).....((((....))))..)))..((.(.((((..((((......((((....)))).....)))).....)))).).)).... ( -45.80) >Sfl_1 480 120 + 1 CAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGCCGCGUGUAUGAAGAAGGCCUUCGGGUUGUAAAGUACUUUCAGCGGGGAGGAAGG ....((((....))))(((((((((.....)))))).....((((....))))..)))..((.(.((((..((((......((((....)))).....)))).....)))).).)).... ( -45.80) >Sty_1 481 120 + 1 CAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGCCGCGUGUAUGAAGAAGGCCUUCGGGUUGUAAAGUACUUUCAGCGGGGAGGAAGG ....((((....))))(((((((((.....)))))).....((((....))))..)))..((.(.((((..((((......((((....)))).....)))).....)))).).)).... ( -45.80) >consensus CAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGCCGCGUGUAUGAAGAAGGCCUUCGGGUUGUAAAGUACUUUCAGCGGGGAGGAAGG ....((((....((..(((((((((.....)))))).....((((....))))..)))..))...((((..((((......((((....)))).....)))).....))))))))..... (-41.70 = -44.70 + 3.00)

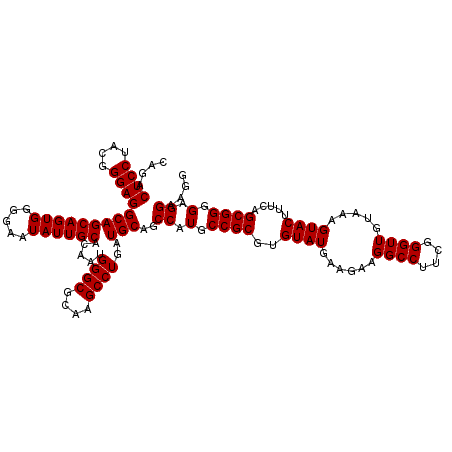

| Location | 4,033,612 – 4,033,732 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -37.76 |
| Energy contribution | -35.77 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4033612 120 + 1 AAGUCGAACGGUAACAGGAAGAAGCUUGCUUCUUUGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGC ........(((((.((((.(((.(((..((..((((((.......))))))..))..)))....)))((....)).))))..............((((((....))))))....))))). ( -38.80) >Hin_1 58 120 + 1 AAGUCGAACGGUAGCAGGAGGAAGCUUGCUUUCUUGCUGACGAGUGGCGGACGGGUGAGUAAUGCUUGGGAAUCUGGCUUAUGGAGGGGGAUAACGACGGGAAACUGUCGCUAAUACCGC ..((((..((.((((((((((........)))))))))).))..))))...(((...(((........(..((((..(((...)))..))))..)(((((....))))))))....))). ( -36.50) >Ype_1 58 120 + 1 AAGUCGAGCGGCAGCGGGAAGUAGUUUACUACUUUGCCGGCGAGCGGCGGACGGGUGAGUAAUGUCUGGGGAUCUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGC ..(((..((.((.((.((.(((((....)))))...)).))..)).)).))).((((..............((((.(((.....))).))))..((((((....))))))....)))).. ( -44.40) >consensus AAGUCGAACGGUAGCAGGAAGAAGCUUGCUUCUUUGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAUCUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGC ..((((..((....((((((((((....))))))).))).))..))))...(((...(((........(..((((.(((.....))).))))..)(((((....))))))))....))). (-37.76 = -35.77 + -1.99)



Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:06:21 2006