
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,426,385 – 3,426,705 |
| Length | 320 |
| Max. P | 0.994367 |

| Location | 3,426,385 – 3,426,505 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -50.07 |
| Consensus MFE | -51.17 |
| Energy contribution | -50.07 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.66 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3426385 120 - 1 GAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGC ..((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......)))))))...(((((...(((((....))))).)))))...... ( -51.20) >Hin_1 41 120 - 1 GAUCUCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCGCAAUGGGGGGAACCCUGACGCAGCCAUGC ..((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......)))))))...(((((...(((((....))))).)))))...... ( -47.80) >Sty_1 41 120 - 1 GAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGC ..((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......)))))))...(((((...(((((....))))).)))))...... ( -51.20) >consensus GAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCCAUGC ..((((..(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......)))))))...(((((...(((((....))))).)))))...... (-51.17 = -50.07 + -1.11)

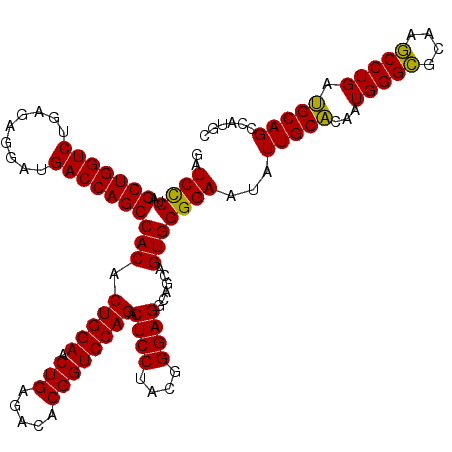

| Location | 3,426,425 – 3,426,545 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -50.53 |
| Consensus MFE | -51.29 |
| Energy contribution | -49.97 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.67 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3426425 120 - 1 GGAUUAGCUUGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGG (((((.(((..((((((((.......)))))))))))...)))))...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))). ( -50.50) >Hin_1 81 120 - 1 GGAUUAGGUAGUUGGUGGGGUAAAUGCCUACCAAGCCUGCGAUCUCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGG (((((((((..((((((((.......))))))))))))..)))))...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))). ( -49.70) >Sty_1 81 120 - 1 GGAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGG (((((.(((..((((((((.......)))))))))))...)))))...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))). ( -51.40) >consensus GGAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGG (((((.(((..((((((((.......)))))))))))...)))))...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))). (-51.29 = -49.97 + -1.33)

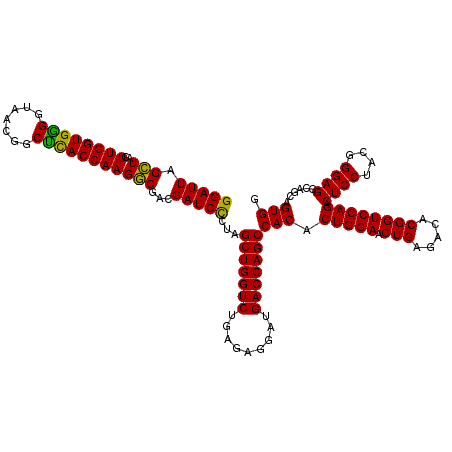

| Location | 3,426,465 – 3,426,585 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -36.70 |
| Energy contribution | -37.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3426465 120 + 1 UCCAGUGUGGCUGGUCAUCCUCUCAGACCAGCUAGGGAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACAAGCUAAUCCCAUCUGGGCACAUCCGAUGGCAAGAGGCCCGAGGGUCCCC .......(((((((((.........)))))))))((((((.(((((((((((...........))))............(((((.(((....))))))))...)))))..)).)))))). ( -45.40) >Hin_1 121 120 + 1 UCCAGUGUGGCUGGUCAUCCUCUCAGACCAGCUAGAGAUCGCAGGCUUGGUAGGCAUUUACCCCACCAACUACCUAAUCCCACUUGGGCUCAUCCUAUGGCAUGCGGCCUCUCAGUCCCG .......(((((((((.........)))))))))(((..((((.(((.(((((................)))))....(((....)))..........))).))))..)))......... ( -37.99) >Sty_1 121 120 + 1 UCCAGUGUGGCUGGUCAUCCUCUCAGACCAGCUAGGGAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUCCCAUCUGGGCACAUCUGAUGGCAAGAGGCCCGAAGGUCCCC .......(((((((((.........)))))))))((((((.((((.((((((((.......))))))))...))...........((((...(((.......))).)))))).)))))). ( -47.70) >consensus UCCAGUGUGGCUGGUCAUCCUCUCAGACCAGCUAGGGAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAUCCCAUCUGGGCACAUCCGAUGGCAAGAGGCCCGAAGGUCCCC (((((..(((((((((.........)))))))))(((((....((.((((((((.......))))))))...))..)))))..)))))...((((...(((.....)))...)))).... (-36.70 = -37.27 + 0.56)



| Location | 3,426,465 – 3,426,585 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -48.83 |
| Consensus MFE | -45.71 |
| Energy contribution | -45.17 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3426465 120 - 1 GGGGACCCUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUUGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGA ((((.((....))))))..(((((...))).))((((..((((((.(((..((((((((.......)))))))))))...))))))..(((((((.........)))))))....)))). ( -48.70) >Hin_1 121 120 - 1 CGGGACUGAGAGGCCGCAUGCCAUAGGAUGAGCCCAAGUGGGAUUAGGUAGUUGGUGGGGUAAAUGCCUACCAAGCCUGCGAUCUCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGA (((.....((((..(((((.((((.((......))..)))).)).((((..((((((((.......)))))))))))))))..)))).(((((((.........)))))))....))).. ( -45.70) >Sty_1 121 120 - 1 GGGGACCUUCGGGCCUCUUGCCAUCAGAUGUGCCCAGAUGGGAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGA ((((.((....))))))......((((.((((.......((((((.(((..((((((((.......)))))))))))...))))))..(((((((.........))))))))))))))). ( -52.10) >consensus GGGGACCCUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUCCCUAGCUGGUCUGAGAGGAUGACCAGCCACACUGGA ((((.((....))))))..(((((...))).))((((..((((((.(((..((((((((.......)))))))))))...))))))..(((((((.........)))))))....)))). (-45.71 = -45.17 + -0.55)

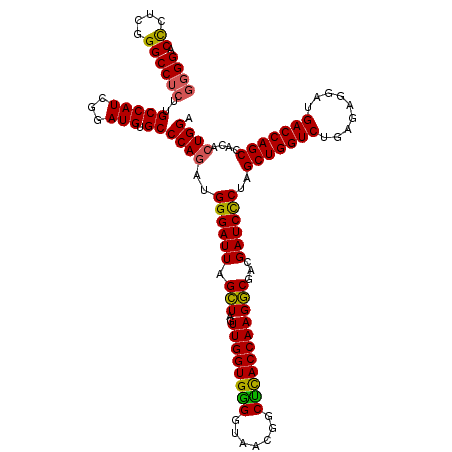
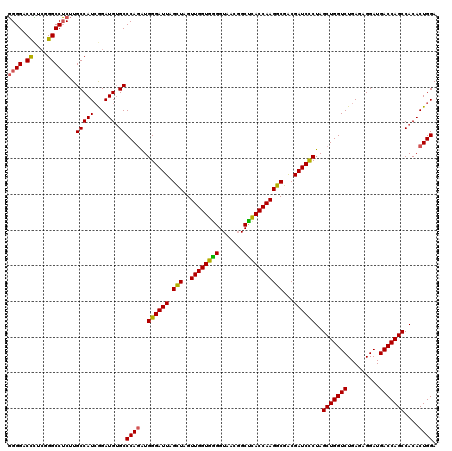
| Location | 3,426,505 – 3,426,625 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -43.83 |
| Consensus MFE | -39.23 |
| Energy contribution | -36.47 |
| Covariance contribution | -2.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3426505 120 - 1 AACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCCUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUUGUUGGUGGGGUAACGGCUCACCAAGGCGAC ..((..(((((((..(((((..((..((((((((...(((((...))))).))..))))))...)).)))))(((....))))))))))(.((((((((.......)))))))).))).. ( -43.90) >Hin_1 161 120 - 1 AACUGUCGCUAAUACCGCGUAUUAUCGGAAGAUGAAAGUGCGGGACUGAGAGGCCGCAUGCCAUAGGAUGAGCCCAAGUGGGAUUAGGUAGUUGGUGGGGUAAAUGCCUACCAAGCCUGC ..((((.((............(((((....)))))..((((((..((....)))))))))).))))......(((....)))..(((((..((((((((.......))))))))))))). ( -41.10) >Sty_1 161 120 - 1 AACGGUGGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCAGAUGUGCCCAGAUGGGAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGAC ...(((((((..((((((.....(((....))).....((((((.((....))))))))))((((((..((.(((....)))....))...)))))).))))..))).))))........ ( -46.50) >consensus AACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCCUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUGGGAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGAC ..((((.((..............(((....)))....((((((..((....)))))))))).))))......(((....)))....(((..((((((((.......)))))))))))... (-39.23 = -36.47 + -2.77)


| Location | 3,426,545 – 3,426,665 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -27.86 |
| Energy contribution | -26.20 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3426545 120 + 1 CAUCUGGGCACAUCCGAUGGCAAGAGGCCCGAGGGUCCCCCUCUUUGGUCUUGCGACGUUAUGCGGUAUUAGCUACCGUUUCCAGUAGUUAUCCCCCUCCAUCAGGCAGUUUCCCAGACA ..((((((.((..(((((((...((((((.(((((...)))))...))))))((.((.....((((((.....)))))).....)).)).........))))).))..))..)))))).. ( -39.90) >Hin_1 201 120 + 1 CACUUGGGCUCAUCCUAUGGCAUGCGGCCUCUCAGUCCCGCACUUUCAUCUUCCGAUAAUACGCGGUAUUAGCGACAGUUUCCCGUCGUUAUCCCCCUCCAUAAGCCAGAUUCCCAAGCA ..((((((...(((...((((.(((((..........)))))............(((((((((.((....(((....))).))))).))))))...........))))))).)))))).. ( -30.70) >Sty_1 201 120 + 1 CAUCUGGGCACAUCUGAUGGCAAGAGGCCCGAAGGUCCCCCUCUUUGGUCUUGCGACGUUAUGCGGUAUUAGCCACCGUUUCCAGUAGUUAUCCCCCUCCAUCAGGCAGUUUCCCAGACA ..((((((.((.(((((((((((((...(((((((......))))))))))))).((.....(((((.......))))).....)).............)))))))..))..)))))).. ( -38.60) >consensus CAUCUGGGCACAUCCGAUGGCAAGAGGCCCGAAGGUCCCCCUCUUUGGUCUUGCGACGUUAUGCGGUAUUAGCCACCGUUUCCAGUAGUUAUCCCCCUCCAUCAGGCAGUUUCCCAGACA ..((((((.((..((((((((....((((....))))..........(((....))).....))((...(((((((........))))))).)).....)))).))..))..)))))).. (-27.86 = -26.20 + -1.66)


| Location | 3,426,545 – 3,426,665 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -54.47 |
| Consensus MFE | -55.14 |
| Energy contribution | -50.60 |
| Covariance contribution | -4.54 |
| Combinations/Pair | 1.32 |
| Mean z-score | -4.94 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3426545 120 - 1 UGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCCUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUG .(((((((..((...(((((((((((((....((((((....))))))......((.......(((....)))....(((((...))))))).)))))).)))))))..)).))))))). ( -55.30) >Hin_1 201 120 - 1 UGCUUGGGAAUCUGGCUUAUGGAGGGGGAUAACGACGGGAAACUGUCGCUAAUACCGCGUAUUAUCGGAAGAUGAAAGUGCGGGACUGAGAGGCCGCAUGCCAUAGGAUGAGCCCAAGUG .(((((((..((...(((((((.(.((..((.((((((....)))))).))...)).)...(((((....)))))..((((((..((....)))))))).)))))))..)).))))))). ( -52.40) >Sty_1 201 120 - 1 UGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUGGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCUUCGGGCCUCUUGCCAUCAGAUGUGCCCAGAUG .(((((((..((...(((((((((((((....((((((....))))))......(((......(((....)))....((((....))))))).)))))).)))))))..)).))))))). ( -55.70) >consensus UGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAGGGGGACCCUCGGGCCUCUUGCCAUCGGAUGUGCCCAGAUG .(((((((..((...(((((((.(.((..((.((((((....)))))).))...)).).....(((....)))....((((((..((....)))))))).)))))))..)).))))))). (-55.14 = -50.60 + -4.54)



| Location | 3,426,585 – 3,426,705 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -35.80 |
| Energy contribution | -34.37 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3426585 120 - 1 AGCUUGCUGUUUCGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAG ..(((((.....((((.......))))((((.(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....)))))))))......... ( -39.20) >Hin_1 241 120 - 1 AGCUUGCUUUCUUGCUGACGAGUGGCGGACGGGUGAGUAAUGCUUGGGAAUCUGGCUUAUGGAGGGGGAUAACGACGGGAAACUGUCGCUAAUACCGCGUAUUAUCGGAAGAUGAAAGUG (((((((((.(((((((........))).)))).)))))).)))..........((((.....(.((..((.((((((....)))))).))...)).)...(((((....))))))))). ( -36.40) >Sty_1 241 120 - 1 AGCUUGCUGCUUCGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUGGCUAAUACCGCAUAACGUCGCAAGACCAAAGAG ..(((((.....((((.......))))((((.(((.(((.(((((((....)).(((......)))))))).((((((....))))))....))))))....)))))))))......... ( -38.90) >consensus AGCUUGCUGUUUCGCUGACGAGUGGCGGACGGGUGAGUAAUGUCUGGGAAACUGCCUGAUGGAGGGGGAUAACUACUGGAAACGGUAGCUAAUACCGCAUAACGUCGCAAGACCAAAGAG .(((..((..((((((.......))))))..))..))).((((..((....((.(((.....))).))....((((((....))))))......))))))...(((....)))....... (-35.80 = -34.37 + -1.43)



Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:05:27 2006