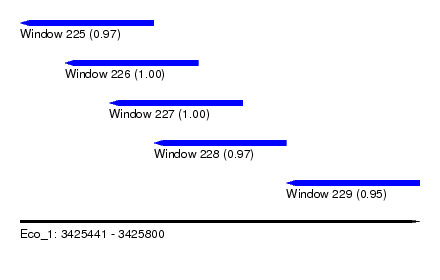
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,425,441 – 3,425,800 |
| Length | 359 |
| Max. P | 0.999891 |

| Location | 3,425,441 – 3,425,561 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -38.90 |
| Energy contribution | -37.18 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.39 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3425441 120 - 1 ACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCUCGCGAGAGCAAGCGGACCUCAUAAAGUGCGUCGUAGUCCGGAUUGGAGUCUGCAACUCGACUCCAUGAAGUCGGAAUCGC ........((....(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...)) ( -39.50) >Hin_1 201 120 - 1 ACACACGUGCUACAAUGGCGUAUACAGAGGGAAGCGAAGCUGCGAGGUGGAGCGAAUCUCAUAAAGUACGUCUAAGUCCGGAUUGGAGUCUGCAACUCGACUCCAUGAAGUCGGAAUCGC ....(((((((....((.......))((((...((...(((....)))...))...))))....))))))).....(((((..(((((((........))))))).....)))))..... ( -34.60) >Sfl_1 201 120 - 1 ACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCUCGCGAGAGCAAGCGGACCUCAUAAAGUGCGUCGUAGUCCGGAUUGGAGUCUGCAACUCGACUCCAUGAAGUCGGAAUCGC ........((....(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...)) ( -39.50) >Sty_1 201 120 - 1 ACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCUCGCGAGAGCAAGCGGACCUCAUAAAGUGCGUCGUAGUCCGGAUUGGAGUCUGCAACUCGACUCCAUGAAGUCGGAAUCGC ........((....(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...)) ( -39.50) >Ype_1 201 120 - 1 ACACACGUGCUACAAUGGCAGAUACAAAGUGAAGCGAACUCGCGAGAGUCAGCGGACCACAUAAAGUCUGUCGUAGUCCGGAUUGGAGUCUGCAACUCGACUCCAUGAAGUCGGAAUCGC ........((....(((((((((.....(((..((..((((....))))..))....))).....)))))))))..(((((..(((((((........))))))).....)))))...)) ( -40.50) >consensus ACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCUCGCGAGAGCAAGCGGACCUCAUAAAGUGCGUCGUAGUCCGGAUUGGAGUCUGCAACUCGACUCCAUGAAGUCGGAAUCGC ........((....(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...)) (-38.90 = -37.18 + -1.72)

| Location | 3,425,481 – 3,425,601 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -43.84 |
| Energy contribution | -42.12 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3425481 120 - 1 GGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCUACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCUCGCGAGAGCAAGCGGACCUCAUAAAGUGCGUCGUAGUCCG ..((((((.....)))))).(((((((.......)))))))....((..((((...(((((((.....(((..((...(((....)))...))....))).....)))))))))))..)) ( -44.20) >Hin_1 241 120 - 1 GGGAUGACGUCAAGUCAUCAUGGCCCUUACGAGUAGGGCUACACACGUGCUACAAUGGCGUAUACAGAGGGAAGCGAAGCUGCGAGGUGGAGCGAAUCUCAUAAAGUACGUCUAAGUCCG ((((((((.....)))))).(((((((.......)))))))...(((((((....((.......))((((...((...(((....)))...))...))))....)))))))......)). ( -37.80) >Sfl_1 241 120 - 1 GGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCUACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCUCGCGAGAGCAAGCGGACCUCAUAAAGUGCGUCGUAGUCCG ..((((((.....)))))).(((((((.......)))))))....((..((((...(((((((.....(((..((...(((....)))...))....))).....)))))))))))..)) ( -44.20) >Sty_1 241 120 - 1 GGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCUACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCUCGCGAGAGCAAGCGGACCUCAUAAAGUGCGUCGUAGUCCG ..((((((.....)))))).(((((((.......)))))))....((..((((...(((((((.....(((..((...(((....)))...))....))).....)))))))))))..)) ( -44.20) >Ype_1 241 120 - 1 GGGAUGACGUCAAGUCAUCAUGGCCCUUACGAGUAGGGCUACACACGUGCUACAAUGGCAGAUACAAAGUGAAGCGAACUCGCGAGAGUCAGCGGACCACAUAAAGUCUGUCGUAGUCCG ..((((((.....)))))).(((((((.......)))))))....((..((((...(((((((.....(((..((..((((....))))..))....))).....)))))))))))..)) ( -45.90) >consensus GGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCUACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCUCGCGAGAGCAAGCGGACCUCAUAAAGUGCGUCGUAGUCCG ..((((((.....)))))).(((((((.......)))))))....((..((((...(((((((.....(((..((...(((....)))...))....))).....)))))))))))..)) (-43.84 = -42.12 + -1.72)

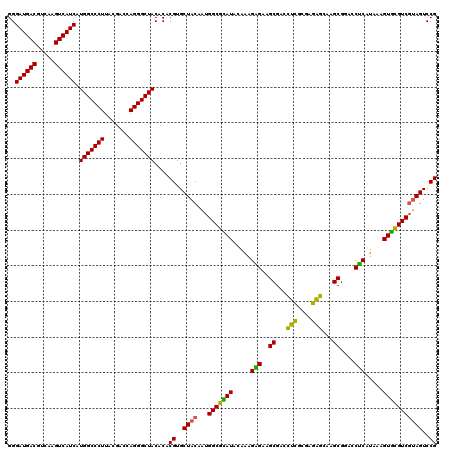
| Location | 3,425,521 – 3,425,641 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -37.56 |
| Energy contribution | -37.08 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3425521 120 - 1 AACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCUACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCU ..(((..((....)).(((((.....)))))......(((..((((((.....)))))).(((((((.......))))))))))..(((((.....))))).......)))......... ( -39.50) >Hin_1 281 120 - 1 AACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGAGUAGGGCUACACACGUGCUACAAUGGCGUAUACAGAGGGAAGCGAAGC ..(((..((....)).(((((.....)))))......(((..((((((.....)))))).(((((((.......))))))))))..(((.(((......)))))).)))........... ( -36.50) >Sfl_1 281 120 - 1 AACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCUACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCU ..(((..((....)).(((((.....)))))......(((..((((((.....)))))).(((((((.......))))))))))..(((((.....))))).......)))......... ( -39.50) >Sty_1 281 120 - 1 AACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCUACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCU ..(((..((....)).(((((.....)))))......(((..((((((.....)))))).(((((((.......))))))))))..(((((.....))))).......)))......... ( -39.50) >Ype_1 281 120 - 1 AACUCAAGGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGAGUAGGGCUACACACGUGCUACAAUGGCAGAUACAAAGUGAAGCGAACU ..(((....))).((((((((.....)))(.((.(..(((..((((((.....)))))).(((((((.......))))))))))...).)).)...)))))................... ( -38.40) >consensus AACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCUACACACGUGCUACAAUGGCGCAUACAAAGAGAAGCGACCU ..(((..((....)).(((((.....)))))......(((..((((((.....)))))).(((((((.......))))))))))..(((((.....))))).......)))......... (-37.56 = -37.08 + -0.48)

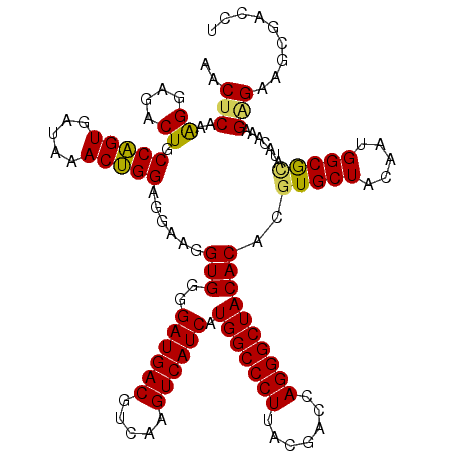
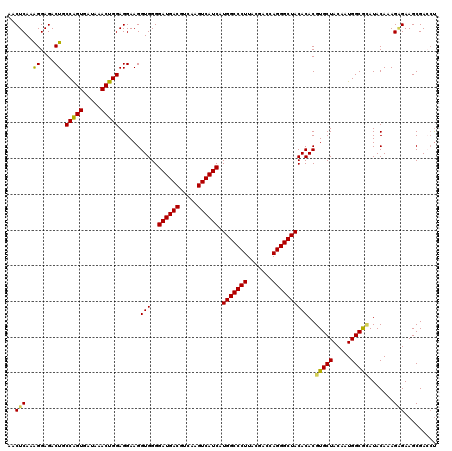
| Location | 3,425,561 – 3,425,680 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -42.40 |
| Energy contribution | -41.96 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3425561 119 - 1 GCGCAACCCUUAUCCUUUGUUGCCAGCG-GUCCGGCCGGGAACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCU ......(((...(((((((...((..((-(.....)))))....)))))))..((.(((((.....))))).)).....)))((((((.....))))))..((((((.......)))))) ( -43.90) >Hin_1 321 119 - 1 GCGCAACCCUUAUCCUUUGUUGCCAGCG-ACUUGGUCGGGAACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGAGUAGGGCU ......(((...(((((((...((..((-((...))))))....)))))))..((.(((((.....))))).)).....)))((((((.....))))))..((((((.......)))))) ( -43.60) >Sfl_1 321 119 - 1 GCGCAACCCUUAUCCUUUGUUGCCAGCG-GUCCGGCCGGGAACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCU ......(((...(((((((...((..((-(.....)))))....)))))))..((.(((((.....))))).)).....)))((((((.....))))))..((((((.......)))))) ( -43.90) >Sty_1 321 119 - 1 GCGCAACCCUUAUCCUUUGUUGCCAGCG-AUUAGGUCGGGAACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCU ......(((....((((((((.((.((.-.....)).)).)))....((....)).(((((.....)))))..))))).)))((((((.....))))))..((((((.......)))))) ( -41.70) >Ype_1 321 120 - 1 GCGCAACCCUUAUCCUUUGUUGCCAGCACGUAAUGGUGGGAACUCAAGGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGAGUAGGGCU ......(((....((((((((.(((.((.....)).))).)))....((....)).(((((.....)))))..))))).)))((((((.....))))))..((((((.......)))))) ( -46.00) >consensus GCGCAACCCUUAUCCUUUGUUGCCAGCG_GUCAGGCCGGGAACUCAAAGGAGACUGCCAGUGAUAAACUGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGACCAGGGCU ......(((....((((((((.((.((.......)).)).)))....((....)).(((((.....)))))..))))).)))((((((.....))))))..((((((.......)))))) (-42.40 = -41.96 + -0.44)
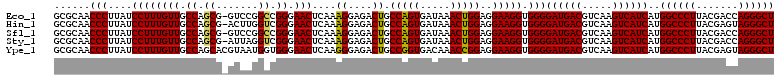

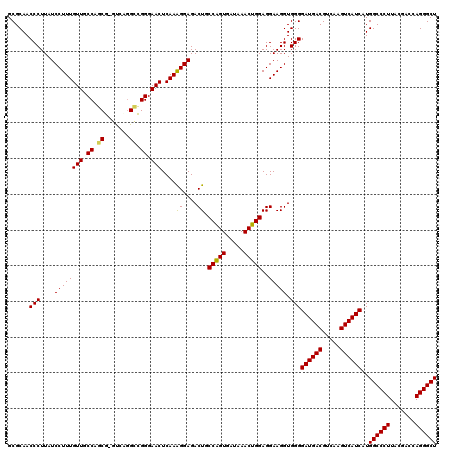
| Location | 3,425,680 – 3,425,800 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -41.98 |
| Energy contribution | -39.90 |
| Covariance contribution | -2.08 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.34 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3425680 120 - 1 UGGUCUUGACAUCCACGGAAGUUUUCAGAGAUGAGAAUGUGCCUUCGGGAACCGUGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUUGUGAAAUGUUGGGUUAAGUCCCGCAACGA .((.((((((...(((((..(((((((....)))))))...((....))..))))).((((..(((.(((((((((....)))).))))).)))...))))..)))))).))........ ( -42.40) >Hin_1 440 120 - 1 UACUCUUGACAUCCUAAGAAGAGCUCAGAGAUGAGCUUGUGCCUUCGGGAACUUAGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUUGUGAAAUGUUGGGUUAAGUCCCGCAACGA ...(((((......))))).(((((((....)))))))((.....(((((.(((((.((((..(((.(((((((((....)))).))))).)))...))))...))))))))))..)).. ( -40.30) >Sfl_1 440 120 - 1 UGGUCUUGACAUCCACGGAAGUUUUCAGAGAUGAGAAUGUGCCUUCGGGAACCGUGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUUGUGAAAUGUUGGGUUAAGUCCCGCAACGA .((.((((((...(((((..(((((((....)))))))...((....))..))))).((((..(((.(((((((((....)))).))))).)))...))))..)))))).))........ ( -42.40) >Sty_1 440 120 - 1 UGGUCUUGACAUCCACAGAACUUUCCAGAGAUGGAUUGGUGCCUUCGGGAACUGUGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUUGUGAAAUGUUGGGUUAAGUCCCGCAACGA .((.((((((...(((((.(((.((((....))))..))).((....))..))))).((((..(((.(((((((((....)))).))))).)))...))))..)))))).))........ ( -41.60) >Ype_1 441 120 - 1 UACUCUUGACAUCCACAGAAUUUGGCAGAGAUGCUAAAGUGCCUUCGGGAACUGUGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUUGUGAAAUGUUGGGUUAAGUCCCGCAACGA ....((((((...(((((..(((((((....)))))))...((....))..))))).((((..(((.(((((((((....)))).))))).)))...))))..))))))........... ( -38.60) >consensus UGGUCUUGACAUCCACAGAAGUUUUCAGAGAUGAGAAUGUGCCUUCGGGAACUGUGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUUGUGAAAUGUUGGGUUAAGUCCCGCAACGA .((.((((((...(((((..(((((((....)))))))...((....))..))))).((((..(((.(((((((((....)))).))))).)))...))))..)))))).))........ (-41.98 = -39.90 + -2.08)
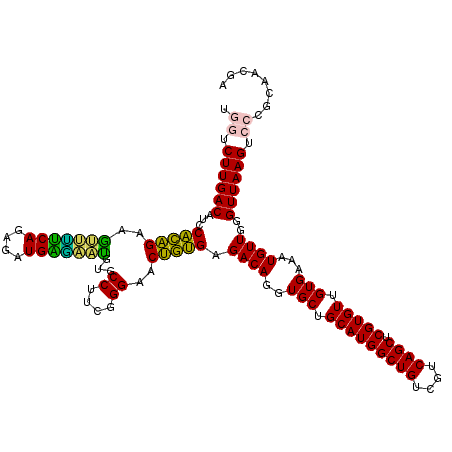
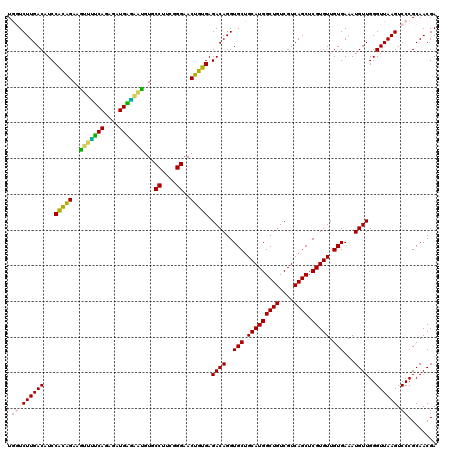
Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:05:19 2006