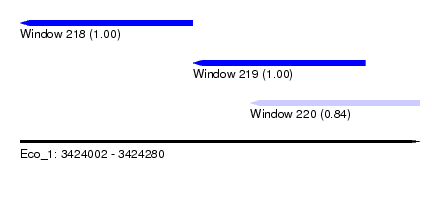
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,424,002 – 3,424,280 |
| Length | 278 |
| Max. P | 0.998520 |

| Location | 3,424,002 – 3,424,122 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -41.59 |
| Energy contribution | -39.27 |
| Covariance contribution | -2.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.44 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3424002 120 - 1 AUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGGCCAAUCAAACCGGGAGAUA ..(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..)))))))((.(....).))................. ( -40.00) >Lpn_1 360 120 - 1 AUCUAGCCAUGUGCAGGAUGAAGGUUGGGUAACACCAACUGGAGGUCCGAACCGGGUAAUGUUGAAAAAUUAUCGGAUGACGUGUGGCUAGGAGUGAAAGGCUAAUCAAGCCCGGAGAUA .((((((((..((((((((...((((((......))))))....))))...((((.((((........)))))))).)).))..)))))))).......((((.....))))........ ( -41.40) >Sty_1 360 120 - 1 AUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACCUGUGGCUGGGGGUGAAAGGCCAAUCAAACCGGGAGAUA ..(((((((((((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)))))))))))((.(....).))................. ( -41.90) >consensus AUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACCUGUGGCUGGGGGUGAAAGGCCAAUCAAACCGGGAGAUA .((((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))))(((.....)))................. (-41.59 = -39.27 + -2.32)

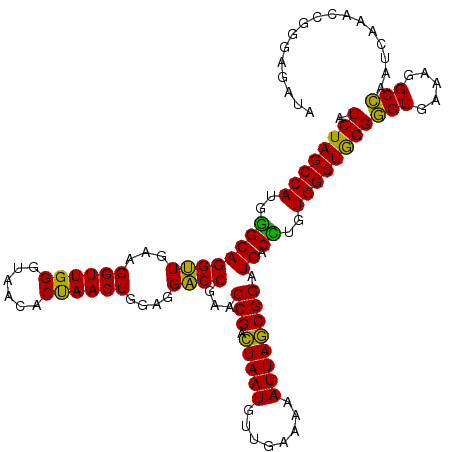
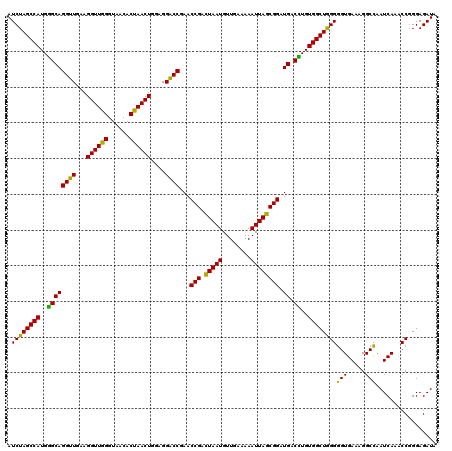
| Location | 3,424,122 – 3,424,242 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -41.94 |
| Energy contribution | -39.28 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.06 |
| Structure conservation index | 1.05 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3424122 120 - 1 CUUUUGUAUAAUGGGUCAGCGACUUAUAUUCUGUAGCAAGGUUAACCGAAUAGGGGAGCCGAAGGGAAACCGAGUCUUAACUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUG ...........(((((...((..((((((((((((((..((((..((.....))..))))...((....))..((((.....)))).....))))))))))))))...))..)))))... ( -41.00) >Lpn_1 480 119 - 1 CUUUUGUAUAAUGGGUCAGCGAGUUACUUUCAGUGGCGAGGUUAACUGAAUAAGGGAGCCGUAGAGAAAUCGAGUCUGAAUAGGGCGAU-AGUCGCUGGGAGUAGACCCGAAACCGGGCG ...........((((((.......((((..(((((((..((((..((.....))..))))...((....))..((((.....))))...-.)))))))..)))))))))).......... ( -38.31) >Sty_1 480 120 - 1 CUUUUGUAUAAUGGGUCAGCGACUUAUAUUCUGUAGCAAGGUUAACCGUAUAGGGGAGCCGUAGGGAAACCGAGUCUUAACCGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUG ...........(((((...((..((((((((((((((..((((..((.....))..))))...((....))..((((.....)))).....))))))))))))))...))..)))))... ( -40.30) >consensus CUUUUGUAUAAUGGGUCAGCGACUUAUAUUCUGUAGCAAGGUUAACCGAAUAGGGGAGCCGUAGGGAAACCGAGUCUUAACAGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUG ...........((((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....)))).....))))))))))))))))))).......... (-41.94 = -39.28 + -2.66)

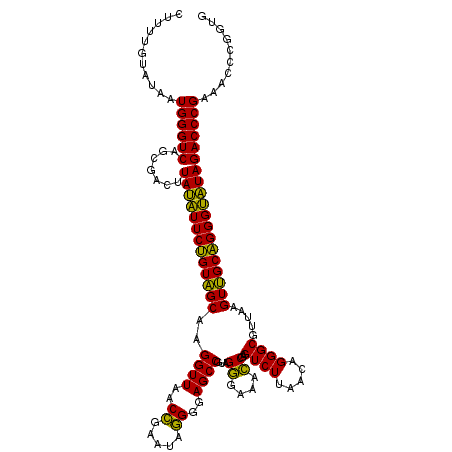
| Location | 3,424,162 – 3,424,280 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -34.44 |
| Energy contribution | -34.33 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3424162 118 - 1 CAAGCAGUGGGAGCAC--GCUUAGGCGUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCUGUAGCAAGGUUAACCGAAUAGGGGAGCCGAAGGGAAACCGAGUCUUAA ...((.(..(..((((--((....))))))..)..)(.((((..........))))).))(((((..............((((..((.....))..))))...((....))))))).... ( -40.60) >Lpn_1 519 120 - 1 CAAGCAGUGGGAGCAUGGUUUAGGCUGUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGAGUUACUUUCAGUGGCGAGGUUAACUGAAUAAGGGAGCCGUAGAGAAAUCGAGUCUGAA ...(((((..((((...))))..)))))((((((((..((((..........))))..)).)))))).(((((...(..((((..((.....))..))))...((....)))...))))) ( -32.50) >Sty_1 520 120 - 1 CAAGCAGUGGGAGCACAGGUUUACCUGUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCUGUAGCAAGGUUAACCGUAUAGGGGAGCCGUAGGGAAACCGAGUCUUAA ...((.(..(..(((((((....)))))))..)..)(.((((..........))))).))(((((..............((((..((.....))..))))...((....))))))).... ( -42.40) >consensus CAAGCAGUGGGAGCAC_GGUUUAGCUGUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCUGUAGCAAGGUUAACCGAAUAGGGGAGCCGUAGGGAAACCGAGUCUUAA ...((.(..(..(((((((....)))))))..)..)(.((((..........))))).))(((((..............((((..((.....))..))))...((....))))))).... (-34.44 = -34.33 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:05:11 2006