
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,422,373 – 3,422,585 |
| Length | 212 |
| Max. P | 0.998469 |

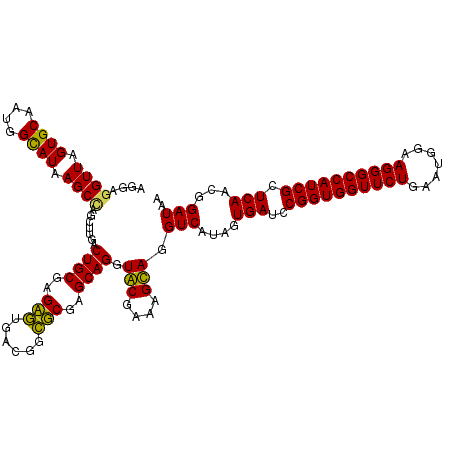
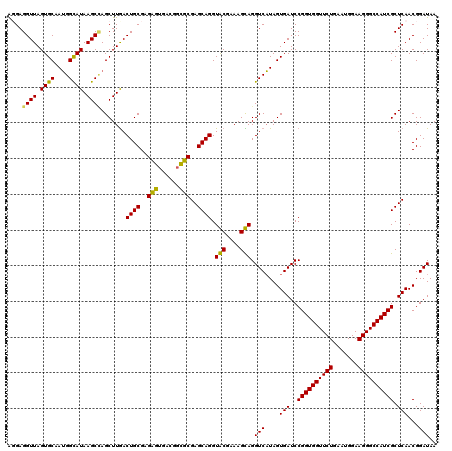
| Location | 3,422,373 – 3,422,493 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -42.92 |
| Consensus MFE | -42.19 |
| Energy contribution | -40.75 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3422373 120 - 1 AGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAGCGUGACGGCGCGAGCAGGUGCGAAAGCAGGUCAUAGUGAUCCGGUGGUUCUGAAUGGAAGGGCCAUCGCUCAACGGAUAA .(((((((.((((....)))).)))).((((((((((..((((....))))..))...(((....)))))))).)))..)))(((((((((.......)))))))))............. ( -46.60) >Hin_1 361 120 - 1 GUGAGGUUAGUGCAAUGGUAUAAGCAAGCUUAACUGCGAGACAGACAAGUCGAGCAGGUACGAAAGUAGGUCAUAGUGAUCCGGUGGUUCUGAAUGGAAGGGCCAUCGCUCAACGGAUAA ((...(((.((((....)))).)))..))....((((..(((......)))..)))).(((....))).(((....(((..((((((((((.......)))))))))).)))...))).. ( -37.20) >Sty_1 361 120 - 1 AGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAGCGUGACGGCGCGAGCAGGUGCGAAAGCAGGUCAUAGUGAUCCGGUGGUUCUGAAUGGAAGGGCCAUCGCUCAACGGAUAA .(((((((.((((....)))).)))).((((((((((..((((....))))..))...(((....)))))))).)))..)))(((((((((.......)))))))))............. ( -46.60) >Ype_1 361 120 - 1 GUGAGGUUAGUGCAAAGGCAUAAGCUAGCUUGACUGCGAGAGUGACGGCUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGAAUGGAAGGGCCAUCGCUCAACGGAUAA ((..((((.((((....)))).)))).))....((((..((((....))))..)))).(((....))).((((...(((..((((((((((.......)))))))))).)))..)))).. ( -41.30) >consensus AGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAGAGUGACGGCGCGAGCAGGUACGAAAGCAGGUCAUAGUGAUCCGGUGGUUCUGAAUGGAAGGGCCAUCGCUCAACGGAUAA ....((((.((((....)))).)))).......((((..(((......)))..)))).(((....))).(((....(((..((((((((((.......)))))))))).)))...))).. (-42.19 = -40.75 + -1.44)

| Location | 3,422,413 – 3,422,533 |
|---|---|
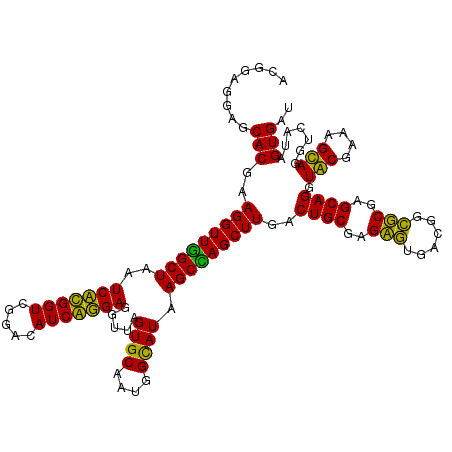
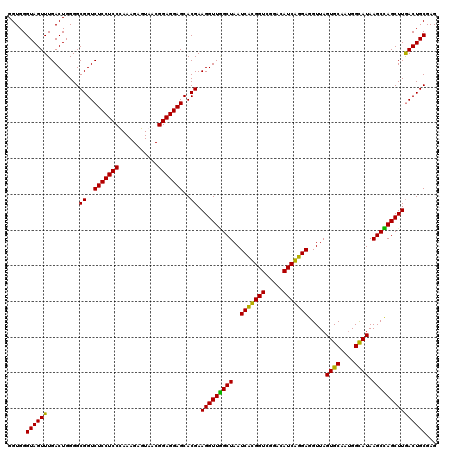
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -44.39 |
| Energy contribution | -41.20 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.49 |
| Structure conservation index | 1.05 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3422413 120 - 1 ACGGAGGAGCACGAAGGUUGGCUAAUCCUGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAGCGUGACGGCGCGAGCAGGUGCGAAAGCAGGUCAUAGUGAU .........(((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))((((((..((((....))))..))...(((....)))))))...))).. ( -45.80) >Hin_1 401 120 - 1 ACGGAGGAGCACGAAGGUUUGCUAAUCACGGUCGGACAUCGUGAGGUUAGUGCAAUGGUAUAAGCAAGCUUAACUGCGAGACAGACAAGUCGAGCAGGUACGAAAGUAGGUCAUAGUGAU .........(((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))).......))).. ( -35.90) >Sty_1 401 120 - 1 ACGGAGGAGCACGAAGGUUGGCUAAUCCUGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAGCGUGACGGCGCGAGCAGGUGCGAAAGCAGGUCAUAGUGAU .........(((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))((((((..((((....))))..))...(((....)))))))...))).. ( -45.80) >Ype_1 401 120 - 1 ACGGAGGAGCACGAAGGUUAGCUAAUCACGGUCGGACAUCGUGAGGUUAGUGCAAAGGCAUAAGCUAGCUUGACUGCGAGAGUGACGGCUCGAGCAGGUACGAAAGUAGGUCUUAGUGAU ((..((((.(....(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..((((....))))..)))).(((....)))).)))).))... ( -41.00) >consensus ACGGAGGAGCACGAAGGUUGGCUAAUCACGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAGAGUGACGGCGCGAGCAGGUACGAAAGCAGGUCAUAGUGAU .........(((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))).......))).. (-44.39 = -41.20 + -3.19)


| Location | 3,422,453 – 3,422,573 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -43.65 |
| Energy contribution | -41.65 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 1.03 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3422453 120 - 1 GGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGCACGAAGGUUGGCUAAUCCUGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAG .(..(...(((((((..(((((..(((((((...........)))))))..))..((....))..)))..))))))).(((((.((((.((((....)))).))))..))))))..)... ( -45.70) >Hin_1 441 120 - 1 GGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGCGUAACGGAGGAGCACGAAGGUUUGCUAAUCACGGUCGGACAUCGUGAGGUUAGUGCAAUGGUAUAAGCAAGCUUAACUGCGAG .....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))... ( -37.90) >Sty_1 441 120 - 1 GGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGCACGAAGGUUGGCUAAUCCUGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAG .(..(...(((((((..(((((..(((((((...........)))))))..))..((....))..)))..))))))).(((((.((((.((((....)))).))))..))))))..)... ( -45.70) >Ype_1 441 120 - 1 GGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGCACGAAGGUUAGCUAAUCACGGUCGGACAUCGUGAGGUUAGUGCAAAGGCAUAAGCUAGCUUGACUGCGAG .....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))... ( -40.70) >consensus GGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGCACGAAGGUUGGCUAAUCACGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAGCUUGACUGCGAG .....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))... (-43.65 = -41.65 + -2.00)


| Location | 3,422,465 – 3,422,585 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -37.39 |
| Energy contribution | -37.83 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3422465 120 - 1 CGGACAGUGUCUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGCACGAAGGUUGGCUAAUCCUGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAG (((((...)))))...(((.(((((((..(((((..(((((((...........)))))))..))..((....))..)))..))))))))))....((((.((((....)))).)))).. ( -43.60) >Hin_1 453 120 - 1 CGGACAGUGUCUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGCGUAACGGAGGAGCACGAAGGUUUGCUAAUCACGGUCGGACAUCGUGAGGUUAGUGCAAUGGUAUAAGCAAG (..((.((((((((((..((((..((((....((..(((((((...........)))))))..))..)))).))))..)))...))))))).))..)(((.((((....)))).)))... ( -37.90) >Sty_1 453 120 - 1 CGGACAGUGUCUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGCACGAAGGUUGGCUAAUCCUGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAG (((((...)))))...(((.(((((((..(((((..(((((((...........)))))))..))..((....))..)))..))))))))))....((((.((((....)))).)))).. ( -43.60) >Ype_1 453 120 - 1 AGGACAGUGUCUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGCACGAAGGUUAGCUAAUCACGGUCGGACAUCGUGAGGUUAGUGCAAAGGCAUAAGCUAG ...((.((((((((((..(((((.((((....((..(((((((...........)))))))..))..)))))))))..)))...))))))).))..((((.((((....)))).)))).. ( -41.00) >consensus CGGACAGUGUCUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGCACGAAGGUUGGCUAAUCACGGUCGGACAUCAGGAGGUUAGUGCAAUGGCAUAAGCCAG (((((...)))))...(((.((((((((((((((..(((((((...........)))))))..))..((....))..)))))))))))))))....((((.((((....)))).)))).. (-37.39 = -37.83 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:05:08 2006