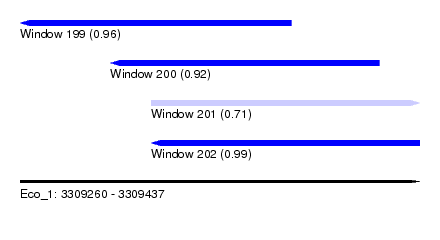
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,309,260 – 3,309,437 |
| Length | 177 |
| Max. P | 0.988095 |

| Location | 3,309,260 – 3,309,380 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -40.42 |
| Energy contribution | -40.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3309260 120 - 1 GCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGCGGCUAAUGAGAGGCUUUACCCACAUAGAGCUGGGUUAGGGUUGUCAUUAGUCGCGAGG (((((..((.(....)....))...))))).......((.((((.......)))).)).((((((((((((.((.(((.(((((........))))).))).)).))))))))))))... ( -41.20) >Sfl_1 1 120 - 1 GCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGCGGCUAAUGAGAGGCUUUACCCACAUAGAGCUGGGUUAGGGUUGUCAUUAGUCGCGAGG (((((..((.(....)....))...))))).......((.((((.......)))).)).((((((((((((.((.(((.(((((........))))).))).)).))))))))))))... ( -41.20) >Sty_1 1 116 - 1 GCAAUUCGCUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGCGGCUAAUGAGAGGCUUUACCCAUAU----CAGGGUUAGGGUUGUCAUUAGUCGCGAGG (((((..((.(....)....))...))))).......((.((((.......)))).)).((((((((((((.((.(((.((((....----..)))).))).)).))))))))))))... ( -40.90) >consensus GCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGCGGCUAAUGAGAGGCUUUACCCACAUAGAGCUGGGUUAGGGUUGUCAUUAGUCGCGAGG (((((..((.(....)....))...))))).......((.((((.......)))).)).((((((((((((.((.(((.(((((........))))).))).)).))))))))))))... (-40.42 = -40.53 + 0.11)

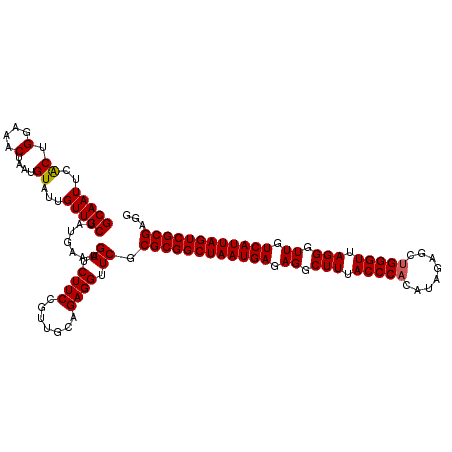
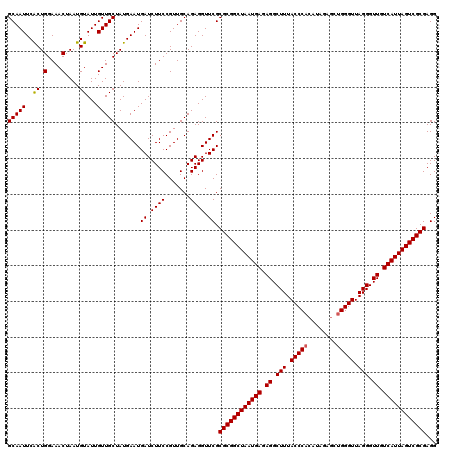
| Location | 3,309,300 – 3,309,419 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -39.77 |
| Energy contribution | -40.00 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3309300 119 - 1 A-AGGGGGCCUGAGUGGCCCCUUUUUUCAAGCUGACGGCAGCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGCGGCUAAUGAGAGGCUUUA (-((((((((.....)))))))))((((((((((.((((((((((.((..(....)...)).)))))))).......((.((((.......)))).)).)))))))..)))))....... ( -42.90) >Sfl_1 41 119 - 1 A-AGGGGGCCUGAGUGGCCCCUUUUUUCAAGCUGACGGCAGCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGCGGCUAAUGAGAGGCUUUA (-((((((((.....)))))))))((((((((((.((((((((((.((..(....)...)).)))))))).......((.((((.......)))).)).)))))))..)))))....... ( -42.90) >Sty_1 37 120 - 1 AGAGGGGGCCUUCAUGGCCCCUUUUUUCCAGCAGAUGGCAGCAAUUCGCUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGCGGCUAAUGAGAGGCUUUA ((((((((((.....))))))))))....(((..(((((((((((.((..(....)...)).)))))))))))......((((((((.((........)))))).....)))).)))... ( -42.00) >consensus A_AGGGGGCCUGAGUGGCCCCUUUUUUCAAGCUGACGGCAGCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGCGGCUAAUGAGAGGCUUUA ..((((((((.....))))))))(((((((((((.((((((((((.((..(....)...)).)))))))).......((.((((.......)))).)).)))))))..))))))...... (-39.77 = -40.00 + 0.23)

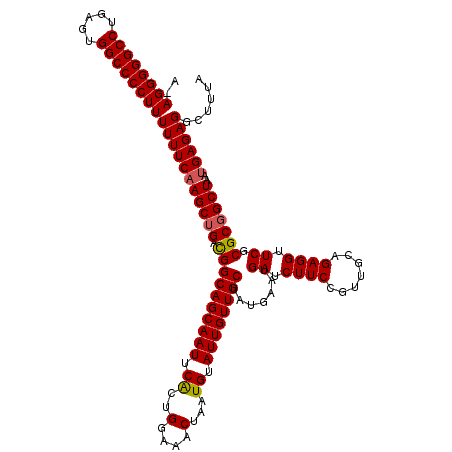
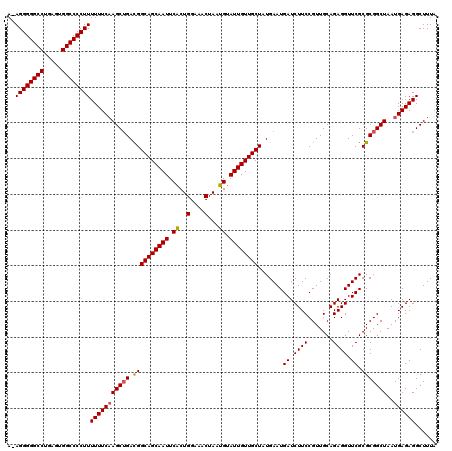
| Location | 3,309,318 – 3,309,437 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.99 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -30.72 |
| Energy contribution | -30.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3309318 119 + 1 GCGCGAACCUCUGCAACGGAAGAUCAUUCAUAGCAACAAUACAUUAGUUUCCAGUGAAUUGCUGCCGUCAGCUUGAAAAAAGGGGCCACUCAGGCCCCCU-UUUCUGAAACUCGCAAGAA ..((((......((.((((.((...((((((.(.(((.........)))..).))))))..)).))))..))..(((((..((((((.....)))))).)-))))......))))..... ( -32.70) >Sfl_1 59 119 + 1 GCGCGAACCUCUGCAACGGAAGAUCAUUCAUAGCAACAAUACAUUAGUUUCCAGUGAAUUGCUGCCGUCAGCUUGAAAAAAGGGGCCACUCAGGCCCCCU-UUUCUGAAACUCGCAAGAA ..((((......((.((((.((...((((((.(.(((.........)))..).))))))..)).))))..))..(((((..((((((.....)))))).)-))))......))))..... ( -32.70) >Sty_1 55 120 + 1 GCGCGAACCUCUGCAACGGAAGAUCAUUCAUAGCAACAAUACAUUAGUUUCCAGCGAAUUGCUGCCAUCUGCUGGAAAAAAGGGGCCAUGAAGGCCCCCUCUUUCUGAAACUCGAAAGAA (((((......)))..(....)..........)).............(((((((((...((....))..)))))))))...((((((.....)))))).((((((........)))))). ( -35.10) >consensus GCGCGAACCUCUGCAACGGAAGAUCAUUCAUAGCAACAAUACAUUAGUUUCCAGUGAAUUGCUGCCGUCAGCUUGAAAAAAGGGGCCACUCAGGCCCCCU_UUUCUGAAACUCGCAAGAA (((((......)))..(....)..........))............(((((((((.....))))..........((((...((((((.....))))))...)))).)))))((....)). (-30.72 = -30.50 + -0.22)

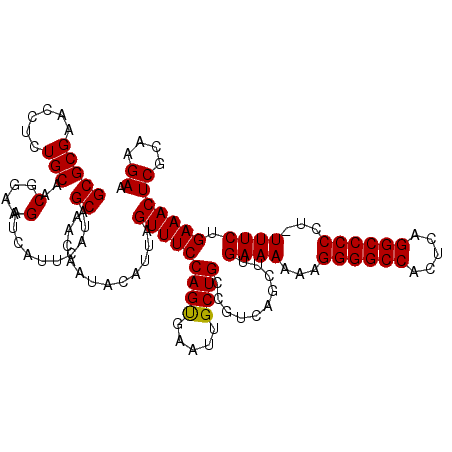

| Location | 3,309,318 – 3,309,437 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.99 |
| Mean single sequence MFE | -47.80 |
| Consensus MFE | -45.67 |
| Energy contribution | -45.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.12 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3309318 119 - 1 UUCUUGCGAGUUUCAGAAA-AGGGGGCCUGAGUGGCCCCUUUUUUCAAGCUGACGGCAGCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGC .....(((((..((.((((-((((((((.....))))))))))))...((.((((((((((((.((..(....)...)).))))))))...(((.....))))))))).))..))))).. ( -47.90) >Sfl_1 59 119 - 1 UUCUUGCGAGUUUCAGAAA-AGGGGGCCUGAGUGGCCCCUUUUUUCAAGCUGACGGCAGCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGC .....(((((..((.((((-((((((((.....))))))))))))...((.((((((((((((.((..(....)...)).))))))))...(((.....))))))))).))..))))).. ( -47.90) >Sty_1 55 120 - 1 UUCUUUCGAGUUUCAGAAAGAGGGGGCCUUCAUGGCCCCUUUUUUCCAGCAGAUGGCAGCAAUUCGCUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGC ......((((..((.(((((((((((((.....)))))))))))))..((..(((((((((((.((..(....)...)).))))))))))).((((......)))))).))..))))... ( -47.60) >consensus UUCUUGCGAGUUUCAGAAA_AGGGGGCCUGAGUGGCCCCUUUUUUCAAGCUGACGGCAGCAAUUCACUGGAAACUAAUGUAUUGUUGCUAUGAAUGAUCUUCCGUUGCAGAGGUUCGCGC ....((((((..((.((((.((((((((.....)))))))).))))..((.((((((((((((.((..(....)...)).))))))))...(((.....))))))))).))..)))))). (-45.67 = -45.57 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:04:54 2006