
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,069,291 – 3,069,419 |
| Length | 128 |
| Max. P | 0.989701 |

| Location | 3,069,291 – 3,069,411 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -29.79 |
| Energy contribution | -31.13 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3069291 120 + 1 CUUAUACUCGUCAUACUUCAAGUUGCAUGUGCUGCGUCUGCGUUCGCUCACCCCAGUCACUUACUUAUGUAAGCUCCUGGGGAUUCACUCUCUUGUCGCCUUCCUGCAACUCGAAUUAUU ............(((.(((.(((((((.(....(((.(.((....))...((((((...(((((....)))))...))))))............).)))...).))))))).))).))). ( -33.20) >Sty_1 1 120 + 1 CUUAUACUCGUCAUACUUCAAGUUGCAUGUGCUGCGGCCGCGUUCCCUCACCCCAGUCACUUACUUUAGUAAGCUCCUGGGGAUUCGCUCGCUUGCCGCCUUCCUGCAACUCGAAUUAUU ............(((.(((.(((((((.(....(((((.(((........((((((...(((((....)))))...)))))).......)))..)))))...).))))))).))).))). ( -40.56) >Ype_1 1 119 + 1 -UUAUACUCGUCAUACUUCAAGCUGCAUAUGCGUUGGCUGCUCUCUUUCACCCGAAUCACUUACUGCAGUAAGCUCAUCGGGACUCUCUCGCUUGCCGCCUUCCUGCAACUCGAAUUAUU -...........(((.(((.((.((((...(.((.(((.((.........(((((....(((((....)))))....)))))........))..))))))....)))).)).))).))). ( -24.63) >consensus CUUAUACUCGUCAUACUUCAAGUUGCAUGUGCUGCGGCUGCGUUCCCUCACCCCAGUCACUUACUUAAGUAAGCUCCUGGGGAUUCACUCGCUUGCCGCCUUCCUGCAACUCGAAUUAUU ............(((.(((.(((((((......(((((.(((........((((((...(((((....)))))...)))))).......)))..))))).....))))))).))).))). (-29.79 = -31.13 + 1.34)



| Location | 3,069,291 – 3,069,411 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -41.01 |
| Energy contribution | -41.68 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.99 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3069291 120 - 1 AAUAAUUCGAGUUGCAGGAAGGCGACAAGAGAGUGAAUCCCCAGGAGCUUACAUAAGUAAGUGACUGGGGUGAGCGAACGCAGACGCAGCACAUGCAACUUGAAGUAUGACGAGUAUAAG .(((.(((((((((((.(...(((..............((((((..((((((....))))))..))))))...((....))...)))..)...))))))))))).)))............ ( -43.80) >Sty_1 1 120 - 1 AAUAAUUCGAGUUGCAGGAAGGCGGCAAGCGAGCGAAUCCCCAGGAGCUUACUAAAGUAAGUGACUGGGGUGAGGGAACGCGGCCGCAGCACAUGCAACUUGAAGUAUGACGAGUAUAAG .(((.(((((((((((.(...(((((..(((..(....((((((..((((((....))))))..)))))).....)..))).)))))..)...))))))))))).)))............ ( -53.10) >Ype_1 1 119 - 1 AAUAAUUCGAGUUGCAGGAAGGCGGCAAGCGAGAGAGUCCCGAUGAGCUUACUGCAGUAAGUGAUUCGGGUGAAAGAGAGCAGCCAACGCAUAUGCAGCUUGAAGUAUGACGAGUAUAA- .(((.(((((((((((.(..(((.((...(....)...(((((...((((((....))))))...))))).........)).)))....)...))))))))))).)))...........- ( -39.40) >consensus AAUAAUUCGAGUUGCAGGAAGGCGGCAAGCGAGAGAAUCCCCAGGAGCUUACUAAAGUAAGUGACUGGGGUGAGAGAACGCAGCCGCAGCACAUGCAACUUGAAGUAUGACGAGUAUAAG .(((.(((((((((((.(...(((((............((((((..((((((....))))))..))))))............)))))..)...))))))))))).)))............ (-41.01 = -41.68 + 0.67)



| Location | 3,069,299 – 3,069,419 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -29.79 |
| Energy contribution | -31.13 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3069299 120 + 1 CGUCAUACUUCAAGUUGCAUGUGCUGCGUCUGCGUUCGCUCACCCCAGUCACUUACUUAUGUAAGCUCCUGGGGAUUCACUCUCUUGUCGCCUUCCUGCAACUCGAAUUAUUUAGAGUAU ....(((.(((.(((((((.(....(((.(.((....))...((((((...(((((....)))))...))))))............).)))...).))))))).))).)))......... ( -33.20) >Sty_1 9 120 + 1 CGUCAUACUUCAAGUUGCAUGUGCUGCGGCCGCGUUCCCUCACCCCAGUCACUUACUUUAGUAAGCUCCUGGGGAUUCGCUCGCUUGCCGCCUUCCUGCAACUCGAAUUAUUUAGAGUAU ....(((.(((.(((((((.(....(((((.(((........((((((...(((((....)))))...)))))).......)))..)))))...).))))))).))).)))......... ( -40.56) >Ype_1 8 120 + 1 CGUCAUACUUCAAGCUGCAUAUGCGUUGGCUGCUCUCUUUCACCCGAAUCACUUACUGCAGUAAGCUCAUCGGGACUCUCUCGCUUGCCGCCUUCCUGCAACUCGAAUUAUUUAGGGUAU ....(((.(((.((.((((...(.((.(((.((.........(((((....(((((....)))))....)))))........))..))))))....)))).)).))).)))......... ( -24.63) >consensus CGUCAUACUUCAAGUUGCAUGUGCUGCGGCUGCGUUCCCUCACCCCAGUCACUUACUUAAGUAAGCUCCUGGGGAUUCACUCGCUUGCCGCCUUCCUGCAACUCGAAUUAUUUAGAGUAU ....(((.(((.(((((((......(((((.(((........((((((...(((((....)))))...)))))).......)))..))))).....))))))).))).)))......... (-29.79 = -31.13 + 1.34)


| Location | 3,069,299 – 3,069,419 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -41.01 |
| Energy contribution | -41.68 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3069299 120 - 1 AUACUCUAAAUAAUUCGAGUUGCAGGAAGGCGACAAGAGAGUGAAUCCCCAGGAGCUUACAUAAGUAAGUGACUGGGGUGAGCGAACGCAGACGCAGCACAUGCAACUUGAAGUAUGACG .........(((.(((((((((((.(...(((..............((((((..((((((....))))))..))))))...((....))...)))..)...))))))))))).))).... ( -43.80) >Sty_1 9 120 - 1 AUACUCUAAAUAAUUCGAGUUGCAGGAAGGCGGCAAGCGAGCGAAUCCCCAGGAGCUUACUAAAGUAAGUGACUGGGGUGAGGGAACGCGGCCGCAGCACAUGCAACUUGAAGUAUGACG .........(((.(((((((((((.(...(((((..(((..(....((((((..((((((....))))))..)))))).....)..))).)))))..)...))))))))))).))).... ( -53.10) >Ype_1 8 120 - 1 AUACCCUAAAUAAUUCGAGUUGCAGGAAGGCGGCAAGCGAGAGAGUCCCGAUGAGCUUACUGCAGUAAGUGAUUCGGGUGAAAGAGAGCAGCCAACGCAUAUGCAGCUUGAAGUAUGACG .........(((.(((((((((((.(..(((.((...(....)...(((((...((((((....))))))...))))).........)).)))....)...))))))))))).))).... ( -39.40) >consensus AUACUCUAAAUAAUUCGAGUUGCAGGAAGGCGGCAAGCGAGAGAAUCCCCAGGAGCUUACUAAAGUAAGUGACUGGGGUGAGAGAACGCAGCCGCAGCACAUGCAACUUGAAGUAUGACG .........(((.(((((((((((.(...(((((............((((((..((((((....))))))..))))))............)))))..)...))))))))))).))).... (-41.01 = -41.68 + 0.67)

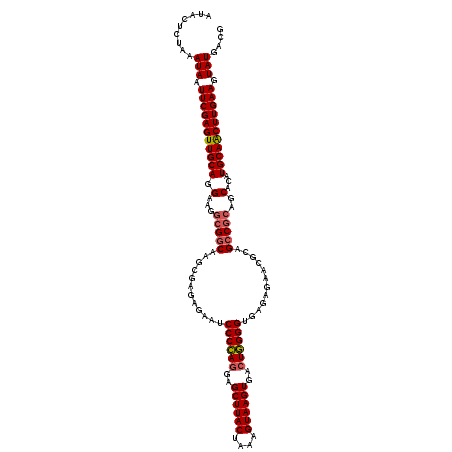
Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:04:47 2006