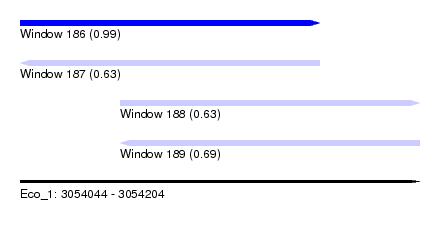
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,054,044 – 3,054,204 |
| Length | 160 |
| Max. P | 0.987959 |

| Location | 3,054,044 – 3,054,164 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -44.69 |
| Energy contribution | -44.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.82 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3054044 120 + 1 CCGAUAUUUCAUACCACAAGAAUGUGGCGCUCCGCGGUUGGUGAGCAUGCUCGGUCCGUCCGAGAAGCCUUAAAACUGCGACGACACAUUCACCUUGAACCAAGGGUUCAAGGGUUACAG ...................(((((((.((...(((((((..((((....(((((.....)))))....)))).))))))).)).))))))).(((((((((...)))))))))....... ( -45.10) >Sfl_1 81 120 + 1 CCGAUAUUUCAUACCACAAGAAUGUGGCGCUCCGCGGUUGGUGAGCAUGCUCGGUUCGUCCGAGAAGCCUUAAAACUGCGACGACACAUUCACCUUGAACCAAGGGUUCAAGGGUUACAG ...................(((((((.((...(((((((..((((....(((((.....)))))....)))).))))))).)).))))))).(((((((((...)))))))))....... ( -45.10) >Sty_1 81 120 + 1 CCGAUAUUUCAUACCACAAGAAUGUGGCGCUCCGCGGUUGGUGAGCAUGCUCGGUUCGUCCGAGAAGCCUUAAAACUGUGACGACACAUUCACCUUGAACCAAGGGUUCAAGGGUUACAG ...................(((((((.((...(((((((..((((....(((((.....)))))....)))).))))))).)).))))))).(((((((((...)))))))))....... ( -43.20) >consensus CCGAUAUUUCAUACCACAAGAAUGUGGCGCUCCGCGGUUGGUGAGCAUGCUCGGUUCGUCCGAGAAGCCUUAAAACUGCGACGACACAUUCACCUUGAACCAAGGGUUCAAGGGUUACAG ...................(((((((.((...(((((((..((((....(((((.....)))))....)))).))))))).)).))))))).(((((((((...)))))))))....... (-44.69 = -44.47 + -0.22)


| Location | 3,054,044 – 3,054,164 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -39.15 |
| Energy contribution | -38.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3054044 120 - 1 CUGUAACCCUUGAACCCUUGGUUCAAGGUGAAUGUGUCGUCGCAGUUUUAAGGCUUCUCGGACGGACCGAGCAUGCUCACCAACCGCGGAGCGCCACAUUCUUGUGGUAUGAAAUAUCGG .......(((((((((...)))))))))(((.((((....))))((((((..(((((.(((..((...(((....))).))..))).)))))((((((....)))))).)))))).))). ( -42.40) >Sfl_1 81 120 - 1 CUGUAACCCUUGAACCCUUGGUUCAAGGUGAAUGUGUCGUCGCAGUUUUAAGGCUUCUCGGACGAACCGAGCAUGCUCACCAACCGCGGAGCGCCACAUUCUUGUGGUAUGAAAUAUCGG (..(((.(((((((((...))))))))).(((((((.((((((.(((....(((..(((((.....)))))...)))....))).)))..))).))))))))))..)............. ( -41.10) >Sty_1 81 120 - 1 CUGUAACCCUUGAACCCUUGGUUCAAGGUGAAUGUGUCGUCACAGUUUUAAGGCUUCUCGGACGAACCGAGCAUGCUCACCAACCGCGGAGCGCCACAUUCUUGUGGUAUGAAAUAUCGG .......(((((((((...)))))))))(((.((((....))))((((((..(((((.(((..(....(((....)))..)..))).)))))((((((....)))))).)))))).))). ( -39.20) >consensus CUGUAACCCUUGAACCCUUGGUUCAAGGUGAAUGUGUCGUCGCAGUUUUAAGGCUUCUCGGACGAACCGAGCAUGCUCACCAACCGCGGAGCGCCACAUUCUUGUGGUAUGAAAUAUCGG .......(((((((((...)))))))))(((.((((....))))((((((..(((((.(((..(....(((....)))..)..))).)))))((((((....)))))).)))))).))). (-39.15 = -38.93 + -0.22)

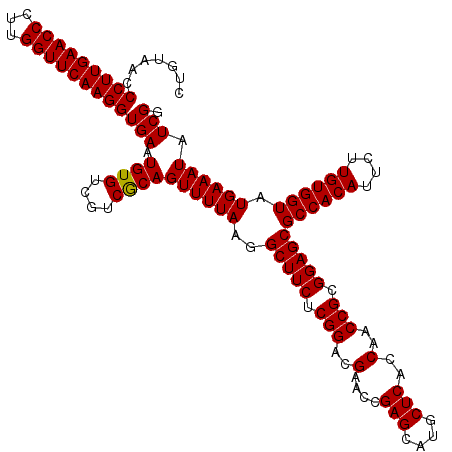
| Location | 3,054,084 – 3,054,204 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -42.87 |
| Consensus MFE | -40.00 |
| Energy contribution | -40.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3054084 120 + 1 GUGAGCAUGCUCGGUCCGUCCGAGAAGCCUUAAAACUGCGACGACACAUUCACCUUGAACCAAGGGUUCAAGGGUUACAGCCUGCGGCGGCAUCUCGGAGAUUCCCUUCUUAUCUGGCAC (((((....)))((....(((((((.(((.......((..((((.....)).(((((((((...)))))))))))..))(((...)))))).)))))))....))...........)).. ( -40.70) >Sfl_1 121 120 + 1 GUGAGCAUGCUCGGUUCGUCCGAGAAGCCUUAAAACUGCGACGACACAUUCACCUUGAACCAAGGGUUCAAGGGUUACAGCCUGCGGCGGCAUCUCGGAGAUUCCCUUCUUAUCUGGCAC (((((....)))((.((.(((((((.(((.......((..((((.....)).(((((((((...)))))))))))..))(((...)))))).)))))))))..))...........)).. ( -41.70) >Sty_1 121 120 + 1 GUGAGCAUGCUCGGUUCGUCCGAGAAGCCUUAAAACUGUGACGACACAUUCACCUUGAACCAAGGGUUCAAGGGUUACAGCCUGCGGCGGCAUCUCGGAGAUUCCCCUUCUGACUGGCGA ((.((((.....((.((.(((((((.(((.......((((((((.....)).(((((((((...)))))))))))))))(((...)))))).)))))))))..)).....)).)).)).. ( -46.20) >consensus GUGAGCAUGCUCGGUUCGUCCGAGAAGCCUUAAAACUGCGACGACACAUUCACCUUGAACCAAGGGUUCAAGGGUUACAGCCUGCGGCGGCAUCUCGGAGAUUCCCUUCUUAUCUGGCAC (((((....)))((....(((((((.(((.......((.(((((.....)).(((((((((...)))))))))))).))(((...)))))).)))))))....))...........)).. (-40.00 = -40.00 + 0.00)

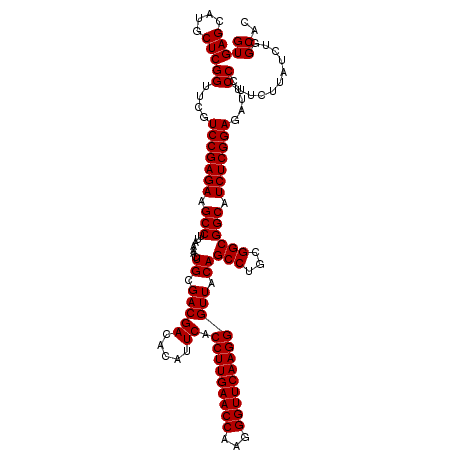
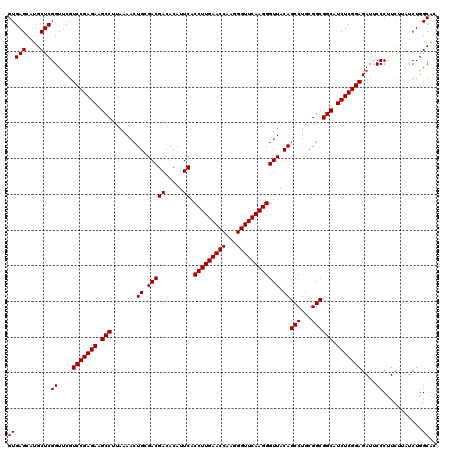
| Location | 3,054,084 – 3,054,204 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -43.07 |
| Energy contribution | -42.63 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3054084 120 - 1 GUGCCAGAUAAGAAGGGAAUCUCCGAGAUGCCGCCGCAGGCUGUAACCCUUGAACCCUUGGUUCAAGGUGAAUGUGUCGUCGCAGUUUUAAGGCUUCUCGGACGGACCGAGCAUGCUCAC ..............((...(((((((((.(((.....((((((..(((((((((((...))))))))).((.....))))..))))))...))).))))))).)).))(((....))).. ( -42.90) >Sfl_1 121 120 - 1 GUGCCAGAUAAGAAGGGAAUCUCCGAGAUGCCGCCGCAGGCUGUAACCCUUGAACCCUUGGUUCAAGGUGAAUGUGUCGUCGCAGUUUUAAGGCUUCUCGGACGAACCGAGCAUGCUCAC ..............((...(((((((((.(((.....((((((..(((((((((((...))))))))).((.....))))..))))))...))).))))))).)).))(((....))).. ( -43.80) >Sty_1 121 120 - 1 UCGCCAGUCAGAAGGGGAAUCUCCGAGAUGCCGCCGCAGGCUGUAACCCUUGAACCCUUGGUUCAAGGUGAAUGUGUCGUCACAGUUUUAAGGCUUCUCGGACGAACCGAGCAUGCUCAC ((.((........)).)).(((((((((.(((.....(((((((.(((((((((((...))))))))).((.....)))).)))))))...))).))))))).))...(((....))).. ( -44.10) >consensus GUGCCAGAUAAGAAGGGAAUCUCCGAGAUGCCGCCGCAGGCUGUAACCCUUGAACCCUUGGUUCAAGGUGAAUGUGUCGUCGCAGUUUUAAGGCUUCUCGGACGAACCGAGCAUGCUCAC ..............((...(((((((((.(((.....(((((((.(((((((((((...))))))))).((.....)))).)))))))...))).))))))).)).))(((....))).. (-43.07 = -42.63 + -0.44)

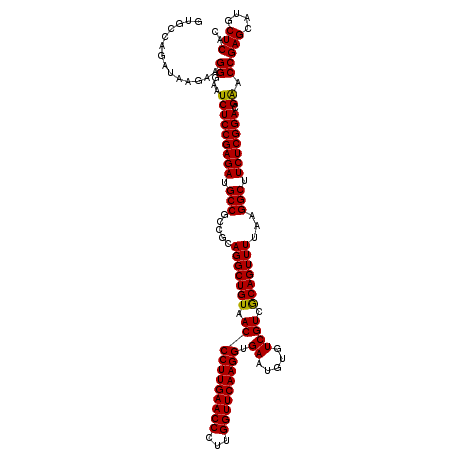
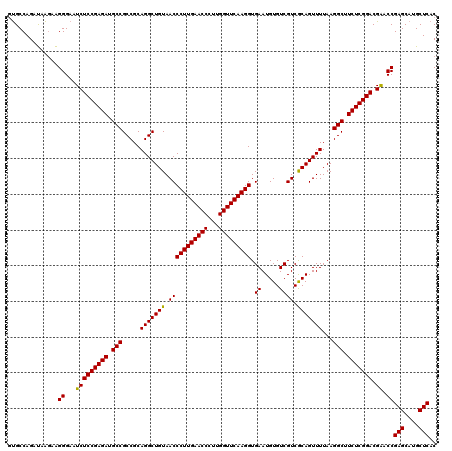
Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:04:43 2006