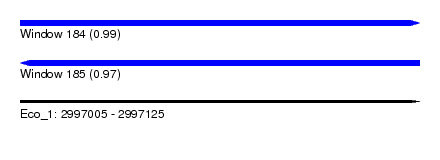
| Sequence ID | Eco_1 |
|---|---|
| Location | 2,997,005 – 2,997,125 |
| Length | 120 |
| Max. P | 0.993898 |

| Location | 2,997,005 – 2,997,125 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -47.61 |
| Consensus MFE | -47.35 |
| Energy contribution | -47.80 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.22 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2997005 120 + 1 UUGGAGCGGGCGAAGGGAAUCGAACCCUCGUAUAGAGCUUGGGAAGCUCUCGUUCUACCAUUGAACUACGCCCGCUUCGAGAUGCGUAAGGCAUUAUAAACCUUACGCUCUCCUUAGCAA ..(((((((((((.(((.......)))))(((.((((((.....)))))).((((.......))))))))))))))))((((.((((((((.........))))))))))))........ ( -51.70) >Sfl_1 1 119 + 1 -UGGAGCGGGCGAAGGGAAUCGAACCCUCGUAUAGAGCUUGGGAAGCUCUCGUUCUACCAUUGAACUACGCCCGCUUCGAGAUGCGUAAGGCAUUAUAAACCUUACGCUCUCCUUAGCAA -.(((((((((((.(((.......)))))(((.((((((.....)))))).((((.......))))))))))))))))((((.((((((((.........))))))))))))........ ( -51.70) >Sty_1 1 110 + 1 -UGGAGCGGGCGAAGGGAAUCGAACCCUCGUAUAGAGCUUGGGAAGCUCUCGUUCUACCAUUGAACUACGCCCGCUUUGAGAUGCGUAAAGCAUUAUAAACCUUACGCUCU--------- -.(((((((((((.(((.......)))))(((.((((((.....)))))).((((.......)))))))))))))))).(((.((((((.............)))))))))--------- ( -39.42) >consensus _UGGAGCGGGCGAAGGGAAUCGAACCCUCGUAUAGAGCUUGGGAAGCUCUCGUUCUACCAUUGAACUACGCCCGCUUCGAGAUGCGUAAGGCAUUAUAAACCUUACGCUCUCCUUAGCAA ..(((((((((((.(((.......)))))(((.((((((.....)))))).((((.......))))))))))))))))((((.((((((((.........))))))))))))........ (-47.35 = -47.80 + 0.45)

| Location | 2,997,005 – 2,997,125 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -51.07 |
| Consensus MFE | -50.25 |
| Energy contribution | -50.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -5.05 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2997005 120 - 1 UUGCUAAGGAGAGCGUAAGGUUUAUAAUGCCUUACGCAUCUCGAAGCGGGCGUAGUUCAAUGGUAGAACGAGAGCUUCCCAAGCUCUAUACGAGGGUUCGAUUCCCUUCGCCCGCUCCAA ........(((((((((((((.......))))))))).))))(.((((((((((((((.......)))).((((((.....)))))).)))(((((........)))))))))))).).. ( -53.80) >Sfl_1 1 119 - 1 UUGCUAAGGAGAGCGUAAGGUUUAUAAUGCCUUACGCAUCUCGAAGCGGGCGUAGUUCAAUGGUAGAACGAGAGCUUCCCAAGCUCUAUACGAGGGUUCGAUUCCCUUCGCCCGCUCCA- ........(((((((((((((.......))))))))).))))(.((((((((((((((.......)))).((((((.....)))))).)))(((((........)))))))))))).).- ( -53.80) >Sty_1 1 110 - 1 ---------AGAGCGUAAGGUUUAUAAUGCUUUACGCAUCUCAAAGCGGGCGUAGUUCAAUGGUAGAACGAGAGCUUCCCAAGCUCUAUACGAGGGUUCGAUUCCCUUCGCCCGCUCCA- ---------((((((((((((.......))))))))).)))...((((((((((((((.......)))).((((((.....)))))).)))(((((........))))))))))))...- ( -45.60) >consensus UUGCUAAGGAGAGCGUAAGGUUUAUAAUGCCUUACGCAUCUCGAAGCGGGCGUAGUUCAAUGGUAGAACGAGAGCUUCCCAAGCUCUAUACGAGGGUUCGAUUCCCUUCGCCCGCUCCA_ ........(((((((((((((.......))))))))).))))..((((((((((((((.......)))).((((((.....)))))).)))(((((........)))))))))))).... (-50.25 = -50.37 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:04:40 2006