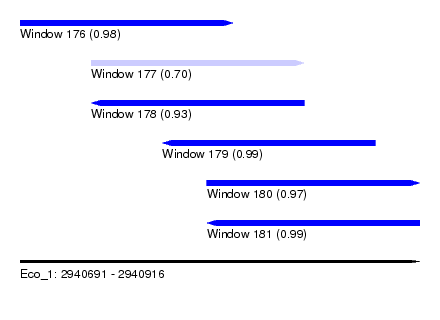
| Sequence ID | Eco_1 |
|---|---|
| Location | 2,940,691 – 2,940,916 |
| Length | 225 |
| Max. P | 0.993266 |

| Location | 2,940,691 – 2,940,811 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -41.01 |
| Energy contribution | -40.13 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2940691 120 + 1 CUACCAGCAAAUACCUAUAGUGGCGGCACUUCCUGAGCCGGAACGAAAAGUUUUAUCGGAAUGCGUGUUCUGGUGAACUUUUGGCUUACGGUUGUGAUGUUGUGUUGUUGUGUUUGCAAU ......((((((((.....(..(((.(((..(((((((((((......(((((.((((((((....)))))))))))))))))))))).))..))).)))..)......))))))))... ( -40.50) >Sfl_1 81 120 + 1 CUACCAGCAAAUACCUAUAGUGGCGGCACUUCCUGAGCCGGAACGAAAAGUUUUAUCGGAAUGCGUAUUCUGGUGAACUUUUGGCUUACGGUUGUGAUGUUGUGUUGUUGUGUUUGCAAU ......((((((((.....(..(((.(((..(((((((((((......(((((.((((((((....)))))))))))))))))))))).))..))).)))..)......))))))))... ( -39.70) >Sty_1 81 120 + 1 CUACCAGCAAAUACCUAUAGUGGCGGCACUUCCUGAGCCGGAACGAAAAGUUUUAUCGGAAUGCGUGUUCUGAUGGGCUUUUGGCUUACGGUUGUGAUGUUGUGUUGUUGUGUUUGCAAU ......((((((((.....(..(((.(((..(((((((((((......(((((.((((((((....)))))))))))))))))))))).))..))).)))..)......))))))))... ( -40.20) >consensus CUACCAGCAAAUACCUAUAGUGGCGGCACUUCCUGAGCCGGAACGAAAAGUUUUAUCGGAAUGCGUGUUCUGGUGAACUUUUGGCUUACGGUUGUGAUGUUGUGUUGUUGUGUUUGCAAU ......((((((((.....(..(((.(((..(((((((((((......(((((.((((((((....)))))))))))))))))))))).))..))).)))..)......))))))))... (-41.01 = -40.13 + -0.88)



| Location | 2,940,731 – 2,940,851 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -34.62 |
| Energy contribution | -33.30 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 1.03 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2940731 120 + 1 GAACGAAAAGUUUUAUCGGAAUGCGUGUUCUGGUGAACUUUUGGCUUACGGUUGUGAUGUUGUGUUGUUGUGUUUGCAAUUGGUCUGCGAUUCAGACCAUGGUAGCAAAGCUACCUUUUU .((((((((((((.((((((((....))))))))))))))))(((((((....)))).))).))))(((((....)))))(((((((.....))))))).((((((...))))))..... ( -34.90) >Sfl_1 121 120 + 1 GAACGAAAAGUUUUAUCGGAAUGCGUAUUCUGGUGAACUUUUGGCUUACGGUUGUGAUGUUGUGUUGUUGUGUUUGCAAUUGGUCUGCGAUUCAGACCAUGGUAGCAAAGCUACCUUUUU .((((((((((((.((((((((....))))))))))))))))(((((((....)))).))).))))(((((....)))))(((((((.....))))))).((((((...))))))..... ( -34.10) >Sty_1 121 120 + 1 GAACGAAAAGUUUUAUCGGAAUGCGUGUUCUGAUGGGCUUUUGGCUUACGGUUGUGAUGUUGUGUUGUUGUGUUUGCAAUUGGUCUGCGAUUCAGACCACGGUAGCGAGACUACCCUUUU .((((((((((((.((((((((....))))))))))))))))(((((((....)))).))).))))(((((....)))))(((((((.....))))))).(((((.....)))))..... ( -32.20) >consensus GAACGAAAAGUUUUAUCGGAAUGCGUGUUCUGGUGAACUUUUGGCUUACGGUUGUGAUGUUGUGUUGUUGUGUUUGCAAUUGGUCUGCGAUUCAGACCAUGGUAGCAAAGCUACCUUUUU ...(.((((((((.((((((((....)))))))))))))))).).....(((.((..(((((....(((((....)))))(((((((.....)))))))...)))))..)).)))..... (-34.62 = -33.30 + -1.32)

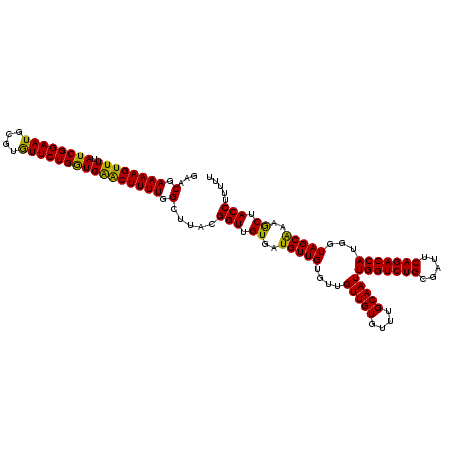
| Location | 2,940,731 – 2,940,851 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2940731 120 - 1 AAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACAACACAACAUCACAACCGUAAGCCAAAAGUUCACCAGAACACGCAUUCCGAUAAAACUUUUCGUUC .....((((((...)))))).(((((((.....)))))))..(((.........................(....)......((((....))))..)))...(((.........)))... ( -25.20) >Sfl_1 121 120 - 1 AAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACAACACAACAUCACAACCGUAAGCCAAAAGUUCACCAGAAUACGCAUUCCGAUAAAACUUUUCGUUC .....((((((...)))))).(((((((.....)))))))..................................(((.(((((((...(.((((....)))).)....))))))).))). ( -24.10) >Sty_1 121 120 - 1 AAAAGGGUAGUCUCGCUACCGUGGUCUGAAUCGCAGACCAAUUGCAAACACAACAACACAACAUCACAACCGUAAGCCAAAAGCCCAUCAGAACACGCAUUCCGAUAAAACUUUUCGUUC .....(((((.....))))).(((((((.....)))))))..................................(((.(((((...(((.(((......))).)))....))))).))). ( -22.00) >consensus AAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACAACACAACAUCACAACCGUAAGCCAAAAGUUCACCAGAACACGCAUUCCGAUAAAACUUUUCGUUC .....((((((...)))))).(((((((.....)))))))..(((.........................(....)......((((....))))..)))...(((.........)))... (-21.84 = -22.07 + 0.23)



| Location | 2,940,771 – 2,940,891 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -30.35 |
| Energy contribution | -30.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.02 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2940771 120 - 1 UAAUCACUAUGGACAGACAGGGUAAAUGUACAGGAAGUGAAAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACAACACAACAUCACAACCGUAAGCCAA .........(((....((...(((....))).((..((((.....((((((...)))))).(((((((.....))))))).......................))))..))))...))). ( -30.70) >Sfl_1 161 120 - 1 UAAUCACUAUGGACAGACAGGGUAAAUGUACAGGAAGUGAAAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACAACACAACAUCACAACCGUAAGCCAA .........(((....((...(((....))).((..((((.....((((((...)))))).(((((((.....))))))).......................))))..))))...))). ( -30.70) >Sty_1 161 120 - 1 UAAUCACUAUGGACAGACAGGGUAAAUGUACAGGAAGUGAAAAAGGGUAGUCUCGCUACCGUGGUCUGAAUCGCAGACCAAUUGCAAACACAACAACACAACAUCACAACCGUAAGCCAA .........(((....((...(((....))).((..((((.....(((((.....))))).(((((((.....))))))).......................))))..))))...))). ( -29.30) >consensus UAAUCACUAUGGACAGACAGGGUAAAUGUACAGGAAGUGAAAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACAACACAACAUCACAACCGUAAGCCAA .........(((....((...(((....))).((..((((.....((((((...)))))).(((((((.....))))))).......................))))..))))...))). (-30.35 = -30.13 + -0.22)


| Location | 2,940,796 – 2,940,916 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -34.80 |
| Energy contribution | -34.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2940796 120 + 1 UGUUGUGUUUGCAAUUGGUCUGCGAUUCAGACCAUGGUAGCAAAGCUACCUUUUUUCACUUCCUGUACAUUUACCCUGUCUGUCCAUAGUGAUUAAUGUAGCACCGCCUAAUUGCGGUGC .(((((....)))))(((((((.....))))))).((((((...)))))).((..(((((...((.(((...........))).)).)))))..))....(((((((......))))))) ( -36.20) >Sfl_1 186 120 + 1 UGUUGUGUUUGCAAUUGGUCUGCGAUUCAGACCAUGGUAGCAAAGCUACCUUUUUUCACUUCCUGUACAUUUACCCUGUCUGUCCAUAGUGAUUAAUGUAGCACCGCCUAAUUGCGGUGC .(((((....)))))(((((((.....))))))).((((((...)))))).((..(((((...((.(((...........))).)).)))))..))....(((((((......))))))) ( -36.20) >Sty_1 186 120 + 1 UGUUGUGUUUGCAAUUGGUCUGCGAUUCAGACCACGGUAGCGAGACUACCCUUUUUCACUUCCUGUACAUUUACCCUGUCUGUCCAUAGUGAUUAAUGUAGCACCGCCAUAUUGCGGUGC .(((((....)))))(((((((.....))))))).(((((.....)))))...............((((((...(((((......)))).)...))))))(((((((......))))))) ( -33.80) >consensus UGUUGUGUUUGCAAUUGGUCUGCGAUUCAGACCAUGGUAGCAAAGCUACCUUUUUUCACUUCCUGUACAUUUACCCUGUCUGUCCAUAGUGAUUAAUGUAGCACCGCCUAAUUGCGGUGC .(((((....)))))(((((((.....))))))).(((((.....)))))...............((((((...(((((......)))).)...))))))(((((((......))))))) (-34.80 = -34.80 + 0.00)


| Location | 2,940,796 – 2,940,916 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -42.05 |
| Energy contribution | -41.83 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.56 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2940796 120 - 1 GCACCGCAAUUAGGCGGUGCUACAUUAAUCACUAUGGACAGACAGGGUAAAUGUACAGGAAGUGAAAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACA (((((((......)))))))((((((.(((....((......)).))).))))))......(((......((((((...)))))).(((((((.....)))))))........))).... ( -42.40) >Sfl_1 186 120 - 1 GCACCGCAAUUAGGCGGUGCUACAUUAAUCACUAUGGACAGACAGGGUAAAUGUACAGGAAGUGAAAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACA (((((((......)))))))((((((.(((....((......)).))).))))))......(((......((((((...)))))).(((((((.....)))))))........))).... ( -42.40) >Sty_1 186 120 - 1 GCACCGCAAUAUGGCGGUGCUACAUUAAUCACUAUGGACAGACAGGGUAAAUGUACAGGAAGUGAAAAAGGGUAGUCUCGCUACCGUGGUCUGAAUCGCAGACCAAUUGCAAACACAACA (((((((......)))))))((((((.(((....((......)).))).))))))......(((......(((((.....))))).(((((((.....)))))))........))).... ( -41.00) >consensus GCACCGCAAUUAGGCGGUGCUACAUUAAUCACUAUGGACAGACAGGGUAAAUGUACAGGAAGUGAAAAAAGGUAGCUUUGCUACCAUGGUCUGAAUCGCAGACCAAUUGCAAACACAACA (((((((......)))))))((((((.(((....((......)).))).))))))......(((......((((((...)))))).(((((((.....)))))))........))).... (-42.05 = -41.83 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:04:37 2006