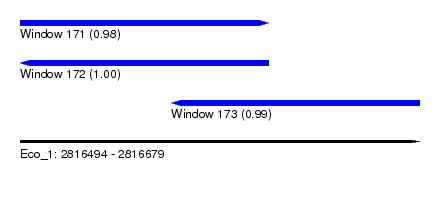
| Sequence ID | Eco_1 |
|---|---|
| Location | 2,816,494 – 2,816,679 |
| Length | 185 |
| Max. P | 0.995544 |

| Location | 2,816,494 – 2,816,609 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -50.43 |
| Consensus MFE | -44.15 |
| Energy contribution | -44.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2816494 115 + 1 AUGGUGCAUCCGGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGGAUGCA-----AAUGGCGGUGAGGCGGGGAUUCGAACCCCGGAUGCA ....((((((((((.(...(((((.(((((.(((((((((.....))))))).(.((..(((..((..(....)...))-----..))).)).).))))))).))))).))))))))))) ( -53.20) >Sfl_1 1 115 + 1 AUGGUGCAUCCGGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGGAUGCA-----AAUGGCGGUGAGGCGGGGAUUCGAACCCCGGAUGCA ....((((((((((.(...(((((.(((((.(((((((((.....))))))).(.((..(((..((..(....)...))-----..))).)).).))))))).))))).))))))))))) ( -53.20) >Sty_1 1 115 + 1 AUGGUGCAUCCGGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGGAUGCA-----AAUGGCGGUGAGGCGGGGAUUCGAACCCCGGAUGCA ....((((((((((.(...(((((.(((((.(((((((((.....))))))).(.((..(((..((..(....)...))-----..))).)).).))))))).))))).))))))))))) ( -53.20) >Ype_1 1 120 + 1 AUGGUGCACCCAGGAAGAUUCGAACUUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGGGUGCAUAGUAAAUGGCGGUGAGGGAGGGAUUCGAACCCUCGAUACA ...((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))...........(((...(((((.......)))))..))). ( -42.10) >consensus AUGGUGCAUCCGGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGGAUGCA_____AAUGGCGGUGAGGCGGGGAUUCGAACCCCGGAUGCA ....((((((((((....((((((.(((((.(((((((((.....)))))))....((((((((...)))..)))))..................))))))).)))))).)))))))))) (-44.15 = -44.65 + 0.50)


| Location | 2,816,494 – 2,816,609 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -47.72 |
| Consensus MFE | -44.59 |
| Energy contribution | -44.65 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2816494 115 - 1 UGCAUCCGGGGUUCGAAUCCCCGCCUCACCGCCAUU-----UGCAUCCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCCGGAUGCACCAU (((((((((((((((((((.(((....(((((...(-----((....(((((((..(((......)))....)))))))...))))))))))).)))))))))...)))))))))).... ( -50.90) >Sfl_1 1 115 - 1 UGCAUCCGGGGUUCGAAUCCCCGCCUCACCGCCAUU-----UGCAUCCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCCGGAUGCACCAU (((((((((((((((((((.(((....(((((...(-----((....(((((((..(((......)))....)))))))...))))))))))).)))))))))...)))))))))).... ( -50.90) >Sty_1 1 115 - 1 UGCAUCCGGGGUUCGAAUCCCCGCCUCACCGCCAUU-----UGCAUCCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCCGGAUGCACCAU (((((((((((((((((((.(((....(((((...(-----((....(((((((..(((......)))....)))))))...))))))))))).)))))))))...)))))))))).... ( -50.90) >Ype_1 1 120 - 1 UGUAUCGAGGGUUCGAAUCCCUCCCUCACCGCCAUUUACUAUGCACCCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAAGUUCGAAUCUUCCUGGGUGCACCAU ......(((((...........)))))..............((((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))).... ( -38.20) >consensus UGCAUCCGGGGUUCGAAUCCCCGCCUCACCGCCAUU_____UGCAUCCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCCGGAUGCACCAU (((((((((((((((((((.(((....(((((............((((..(((...))))))).......((((.......)))))))))))).)))))))))...)))))))))).... (-44.59 = -44.65 + 0.06)

| Location | 2,816,564 – 2,816,679 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -51.88 |
| Consensus MFE | -47.55 |
| Energy contribution | -46.68 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 2816564 115 - 1 AAGAGGCGUGUGGUGAGGUGGCCGAGAGGCUGAAGGCGCUCCCCUGCUAAGGGAGUAUGCGGUCAAAAGCUGCAUCCGGGGUUCGAAUCCCCGCCUCACCGCCAUU-----UGCAUCCGU ..((.(((.((((((((((((((....))))...((.((((((.......))))))(((((((.....)))))))))((((.......))))))))))))))....-----))).))... ( -53.70) >Sfl_1 71 115 - 1 AAGAGGCGUGUGGUGAGGUGGCCGAGAGGCUGAAGGCGCUCCCCUGCUAAGGGAGUAUGCGGUCAAAAGCUGCAUCCGGGGUUCGAAUCCCCGCCUCACCGCCAUU-----UGCAUCCGU ..((.(((.((((((((((((((....))))...((.((((((.......))))))(((((((.....)))))))))((((.......))))))))))))))....-----))).))... ( -53.70) >Sty_1 71 115 - 1 AAGAGGCGUGUGGUGAGGUGGCCGAGAGGCUGAAGGCGCUCCCCUGCUAAGGGAGUAUGCGGUCAAAAGCUGCAUCCGGGGUUCGAAUCCCCGCCUCACCGCCAUU-----UGCAUCCGU ..((.(((.((((((((((((((....))))...((.((((((.......))))))(((((((.....)))))))))((((.......))))))))))))))....-----))).))... ( -53.70) >Ype_1 71 120 - 1 AAGAAGCGUAUGGUGAGGUGGCCGAGAGGCUGAAGGCGCUCCCCUGCUAAGGGAGUAUACGGUCAAAAGCUGUAUCGAGGGUUCGAAUCCCUCCCUCACCGCCAUUUACUAUGCACCCAU .......((((((((((..((((....))))...(((((((((.......))))))(((((((.....))))))).(((((...........)))))...))).))))))))))...... ( -46.40) >consensus AAGAGGCGUGUGGUGAGGUGGCCGAGAGGCUGAAGGCGCUCCCCUGCUAAGGGAGUAUGCGGUCAAAAGCUGCAUCCGGGGUUCGAAUCCCCGCCUCACCGCCAUU_____UGCAUCCGU .......(.(((((((((.((((....))))......((((((.......))))))(((((((.....))))))).(((((.......)))))))))))))))................. (-47.55 = -46.68 + -0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:04:30 2006