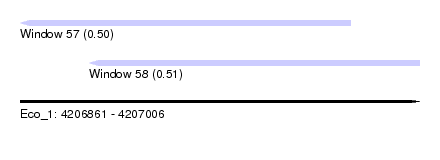
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,206,861 – 4,207,006 |
| Length | 145 |
| Max. P | 0.510158 |

| Location | 4,206,861 – 4,206,981 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -31.13 |
| Energy contribution | -32.13 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>Eco_1 4206861 120 - 1 GCUAAAUCGCAUCCCUCUAUGCCGUACCGUUGUGGCCCAUGUUUGGCCACGACCCGCUCGGCGCCGGUCGACCUUCCUCUGUCGGUCCUUUACGACGAAGCUUGUGACCACGUUUACAUC ((......))..........((((....(((((((((.......))))))))).....))))...(((((..((((....((((........))))))))....)))))........... ( -38.10) >Hin_1 201 120 - 1 UCUAAAUCGCAUCCCUCUAUGCCGUACCGUUGUGGCCCAUGUUUGGCCACGUCCCGCUCGGCGUAAACAGACUUUCCUUUGUCGGUCCACUACGACGAAGCUUCUGACCACGUUUACAUC ..((((.((........(((((((...((.(((((((.......)))))))...))..)))))))..((((.....((((((((........))))))))..))))....)))))).... ( -35.90) >Sty_1 201 120 - 1 GCUAAAUCGCAUCCCUCUAUGCCGUACCGUUGUGGCCCAUGUUUGGCCACGACCCGCUCGGCGUCGGUCGACCUUCCUCUGUCGGUCCUUUACGACGAAGCUUGUGACCACGUUUACAUC ((......))........((((((....(((((((((.......))))))))).....)))))).(((((..((((....((((........))))))))....)))))........... ( -39.00) >consensus GCUAAAUCGCAUCCCUCUAUGCCGUACCGUUGUGGCCCAUGUUUGGCCACGACCCGCUCGGCGUCGGUCGACCUUCCUCUGUCGGUCCUUUACGACGAAGCUUGUGACCACGUUUACAUC ......(((((..(.((.((((((....(((((((((.......))))))))).....))))))................((((........)))))).)..)))))............. (-31.13 = -32.13 + 1.00)



| Location | 4,206,886 – 4,207,006 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -34.22 |
| Energy contribution | -33.23 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4206886 120 - 1 CGGAUGGGAGGUUGUUCCAGCGUGAUCGGUAAUCCGUGCCACUCCGCCCUACCCCGUCGUCCGUUCUCGGCGCGACCUUGGAACCACUUCCGGCCCUAACUAACCUCGGCUACUUAAAUU ......((((((.(((((((((.((..((((.....))))..)))))........((((((((....))).)))))..)))))).))))))((((............))))......... ( -39.50) >Hin_1 226 120 - 1 CGGAUAGGAGAUGGUUCUAGCGUGACCGGUAAUCCGUGCCACUCCGCCCUACCCCGUCGUCCGUUCUCGGCGCGACAUUGGAACCACUCCUAUUCCUUACUAAACUCGGCUACACAAAUU .(((((((((.(((((((((((.((..((((.....))))..)))))........((((((((....))).)))))..)))))))))))))))))......................... ( -43.40) >Sty_1 226 120 - 1 CGGAUGGGAGGUUGUUCCAGCGUGAUCGGUAAUCCGUGCCACUCCGCCCUACCCCGUCGUCCGUUCUCGGCGCGACCUUGGAACCACUUAUGGCCCUAACUAACCUCGGCUACUUAAAUU ....((((((((.(((((((((.((..((((.....))))..)))))........((((((((....))).)))))..)))))).)))).(((((............))))))))).... ( -34.70) >consensus CGGAUGGGAGGUUGUUCCAGCGUGAUCGGUAAUCCGUGCCACUCCGCCCUACCCCGUCGUCCGUUCUCGGCGCGACCUUGGAACCACUUCUGGCCCUAACUAACCUCGGCUACUUAAAUU .((.((((((...(((((((((.((..((((.....))))..)))))........((((((((....))).)))))..))))))..)))))).))......................... (-34.22 = -33.23 + -0.99)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Dec 7 14:12:58 2006