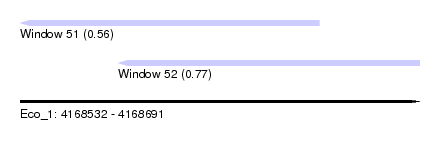
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,168,532 – 4,168,691 |
| Length | 159 |
| Max. P | 0.765233 |

| Location | 4,168,532 – 4,168,651 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -38.74 |
| Energy contribution | -38.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4168532 119 - 1 UUAUUUAAGAGGAUUCCUUGACUGGCAGAAUCCUCCUCGGCUGGAGGAGCGCCAUGCCGCUACCUGUACUCACUGUCUCAUGACGUGGGUACGCAGACCGUACAUAGGUUUCGC-UCCAC ........(((((..(((((((.((((...((((((......))))))......)))).......((((((((.(((....))))))))))).......)).)).)))..)).)-))... ( -42.10) >Hin_1 120 119 - 1 UUAUUUAAGAGGGUUCCUUGACUGGCAGAAUCCUCCUCGGCUGGAGGAGCGCCAUGCCGCUACCUGUACUCACUGUCUCAUGACGUGGGUAUGCAGACCGUACAUUGUUUUCGU-UCCAC ........((((((((((.....))..))))))))..((((((.(((((((......)))).)))((((((((.(((....))))))))))).))).)))..............-..... ( -37.20) >Ype_1 121 120 - 1 UUAUUUAAGAGGAUUCCUUGACUGGCAGAAUCCUCCUCGGCUGGAGGAGCGCCAUGCCGCUACCUGUACUCACUGUCUCAUGACGUGGGUAUGCAGACCGUACAUAGGUUUCGCCUCCAC ........((((...(((((((.((((...((((((......))))))......)))).....((((((((((.(((....)))))))))..))))...)).)).))).....))))... ( -41.10) >consensus UUAUUUAAGAGGAUUCCUUGACUGGCAGAAUCCUCCUCGGCUGGAGGAGCGCCAUGCCGCUACCUGUACUCACUGUCUCAUGACGUGGGUAUGCAGACCGUACAUAGGUUUCGC_UCCAC ........((((((((((.....))..))))))))..((((((.(((((((......)))).)))((((((((.(((....))))))))))).))).))).................... (-38.74 = -38.30 + -0.44)


| Location | 4,168,571 – 4,168,691 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -39.67 |
| Energy contribution | -39.67 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4168571 120 - 1 GGCCUAAAUGGGCUCCCAUCUGCUAGUGAAGAGCCCUAUUUCACGGCUCGGACACACCGAUGUCGUCUGAAAAUAUGCAUCGCUACUCGCGUAAGGUGCUCAAGAAUUAGUUGGGCGCUU .(((((.((((....))))..((((((...(((((((.(((((((((((((.....)))).)))))..)))).(((((..........)))))))).))))....))))))))))).... ( -40.30) >Hin_1 159 120 - 1 GUCCUAGAUGGGCUCCCAUCUGCUAGUGAAGAGCCCUAUUUCACGGCUCGGAUACACCGAUGUCGUCUGAAAAUAUGCAUCGCUACUCGCGUAAGGUGCUCAAGAAUUAGUUGGGCGCUU ((((.((((((....))))))((((((...(((((((.(((((((((((((.....)))).)))))..)))).(((((..........)))))))).))))....)))))).)))).... ( -43.50) >Ype_1 161 120 - 1 GGCCUAAAUGGGCUCCCAUCUGCUAGUGAAGAGCCCUAUUUCACGGCUCGGACACACCGAUGUCGUCUGAAAAUAUGCAUCGCUACUCGCGUAAGGUGCUCAAGAAUUAGUUGGGCGCUU .(((((.((((....))))..((((((...(((((((.(((((((((((((.....)))).)))))..)))).(((((..........)))))))).))))....))))))))))).... ( -40.30) >consensus GGCCUAAAUGGGCUCCCAUCUGCUAGUGAAGAGCCCUAUUUCACGGCUCGGACACACCGAUGUCGUCUGAAAAUAUGCAUCGCUACUCGCGUAAGGUGCUCAAGAAUUAGUUGGGCGCUU ..((.....))((.((((...((((((...(((((((.(((((((((((((.....)))).)))))..)))).(((((..........)))))))).))))....)))))))))).)).. (-39.67 = -39.67 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Dec 7 14:12:38 2006