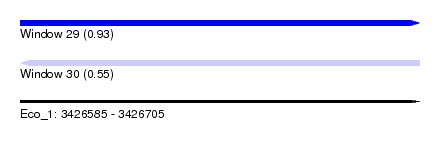
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,426,585 – 3,426,705 |
| Length | 120 |
| Max. P | 0.933681 |

| Location | 3,426,585 – 3,426,705 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -27.21 |
| Energy contribution | -27.99 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
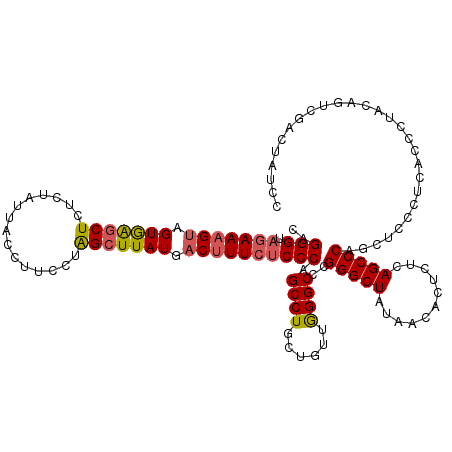
>Eco_1 3426585 120 + 1 CAGGGUAGAAAGUAGUGAGCUCUCUAUUACCUUUCUAGCUUAUGACUUUCUCCCAGCCUGCUGUUGGGCCCGGGCUAUAACACUCUCAGCCCAGCUCCCUCACCCUACAGUCGACUAUCC ..(((.(((((((.(((((((...............))))))).))))))))))((.(.(((((.(((...(((((...........)))))((....))..))).))))).).)).... ( -37.06) >Hin_1 241 120 + 1 CCGGGUCCAAAGAAGUAGACUUUCUAUUACCUUGCUGGCUUAUCACAUUCUCCCAGCCUGCUCUUAGGCACGGGCUACAACACUCUCAGCCCAGCUCCCUCACCCUAAAAAAGACUAUCC ..((((((.(((((((((.....))))).....((.((((..............)))).)))))).))...(((((...........))))).........))))............... ( -25.94) >Sty_1 241 120 + 1 CAGGGUAGAAAGUAGUGAGCUCUCUAUUACCUUCCUAGCUUAUGACUUUCUCCCAGCCUGCUGUUGGGCCCGGGCUGUAACACUCUCAGCCCAGCUCCCUCACCCUACAGUCGACUAUCC ..(((.(((((((.(((((((...............))))))).))))))))))((.(.(((((.(((...((((((.........))))))((....))..))).))))).).)).... ( -40.56) >consensus CAGGGUAGAAAGUAGUGAGCUCUCUAUUACCUUCCUAGCUUAUGACUUUCUCCCAGCCUGCUGUUGGGCCCGGGCUAUAACACUCUCAGCCCAGCUCCCUCACCCUACAGUCGACUAUCC ..(((.(((((((.(((((((...............))))))).)))))))))).((((......))))..(((((...........)))))............................ (-27.21 = -27.99 + 0.79)

| Location | 3,426,585 – 3,426,705 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -40.28 |
| Consensus MFE | -33.74 |
| Energy contribution | -32.87 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
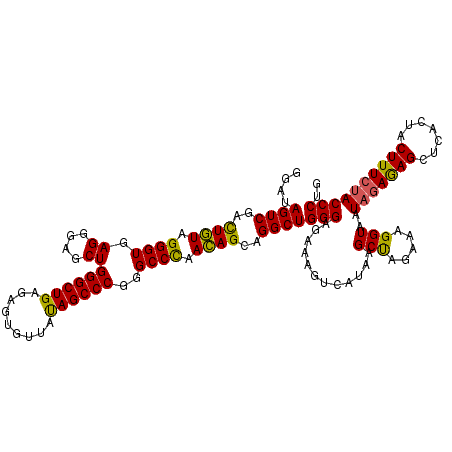
>Eco_1 3426585 120 - 1 GGAUAGUCGACUGUAGGGUGAGGGAGCUGGGCUGAGAGUGUUAUAGCCCGGGCCCAACAGCAGGCUGGGAGAAAGUCAUAAGCUAGAAAGGUAAUAGAGAGCUCACUACUUUCUACCCUG ....((((..((((.((((.......((((((((.........)))))))))))).))))..))))((((((((((....((((...............))))....))))))).))).. ( -40.77) >Hin_1 241 120 - 1 GGAUAGUCUUUUUUAGGGUGAGGGAGCUGGGCUGAGAGUGUUGUAGCCCGUGCCUAAGAGCAGGCUGGGAGAAUGUGAUAAGCCAGCAAGGUAAUAGAAAGUCUACUUCUUUGGACCCGG ...............(((((((((((((..((((.(..(((..((.((((.((((......))))))))....))..)))..)))))..))...(((.....))))))))))..)))).. ( -36.90) >Sty_1 241 120 - 1 GGAUAGUCGACUGUAGGGUGAGGGAGCUGGGCUGAGAGUGUUACAGCCCGGGCCCAACAGCAGGCUGGGAGAAAGUCAUAAGCUAGGAAGGUAAUAGAGAGCUCACUACUUUCUACCCUG ....((((..((((.((((.......((((((((.........)))))))))))).))))..))))((((((((((....((((...............))))....))))))).))).. ( -43.17) >consensus GGAUAGUCGACUGUAGGGUGAGGGAGCUGGGCUGAGAGUGUUAUAGCCCGGGCCCAACAGCAGGCUGGGAGAAAGUCAUAAGCUAGAAAGGUAAUAGAGAGCUCACUACUUUCUACCCUG ....((((..((((.((((.((....))((((((.........))))))..)))).))))..))))(((............(((.....)))..(((((((.......)))))))))).. (-33.74 = -32.87 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Dec 7 14:11:28 2006