
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,604,312 – 4,604,472 |
| Length | 160 |
| Max. P | 0.864560 |

| Location | 4,604,312 – 4,604,432 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -40.13 |
| Energy contribution | -42.47 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4604312 120 + 1 AGUGCGAGCAAUAUCAACGUGGUUUUUGGUGCGAGGGGGGGGACUUGAACCCCCACGUCCGUAAGGACACUAACACCUGAAGCUAGCGCGUCUACCAAUUCCGCCACCUUCGCAUACCAU .(((((((...........(((((((.((((..((.(((((........)))))..((((....)))).))..)))).)))))))(.(((...........))))...)))))))..... ( -44.00) >Sfl_1 241 120 + 1 AGUGCGAGCAAUAUCAACGUGAUUUUUGGUGCGAGGGGGGGGACUUGAACCCCCACGUCCGUAAGGACACUAACACCUGAAGCUAGCGCGUCUACCAAUUCCGCCACCUUCGCAUACCAU .(((((((....(((.....)))....((((((((((((((........)))))..((((....)))).......)))((.((....)).)).........)).)))))))))))..... ( -40.50) >Sty_1 192 119 + 1 AGUGCGAGCAAUAUCAACGUGGUUUU-GGUGCGAGGGGGGGGACUUGAACCCCCACGUCCGUAAGGACACUAACACCUGAAGCUAGCGCGUCUACCAAUUCCGCCACCUUCGCAUACCAU .(((((((..........((((..((-((((..((((((((........)))))..((((....)))).......)))((.((....)).))))))))..))))....)))))))..... ( -43.54) >consensus AGUGCGAGCAAUAUCAACGUGGUUUUUGGUGCGAGGGGGGGGACUUGAACCCCCACGUCCGUAAGGACACUAACACCUGAAGCUAGCGCGUCUACCAAUUCCGCCACCUUCGCAUACCAU .(((((((...........(((((((.((((..((.(((((........)))))..((((....)))).))..)))).)))))))(.(((...........))))...)))))))..... (-40.13 = -42.47 + 2.33)


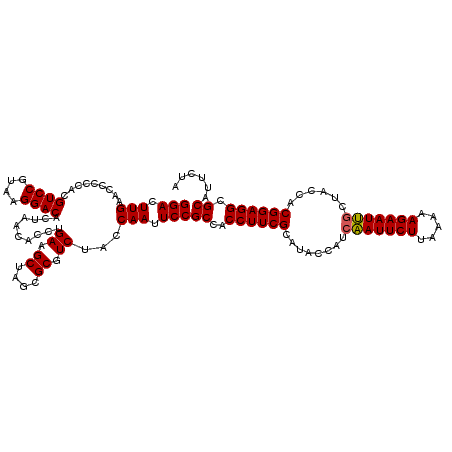
| Location | 4,604,352 – 4,604,472 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -29.59 |
| Consensus MFE | -27.45 |
| Energy contribution | -29.34 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4604352 120 + 1 GGACUUGAACCCCCACGUCCGUAAGGACACUAACACCUGAAGCUAGCGCGUCUACCAAUUCCGCCACCUUCGCAUACCAUCAAUUCUUAAAAAGAAUUGCUACCACGGAGGCGCAUUCUA (((.(((.........((((....))))..........((.((....)).))...))).)))((..((((((........(((((((.....)))))))......)))))).))...... ( -29.84) >Sfl_1 281 120 + 1 GGACUUGAACCCCCACGUCCGUAAGGACACUAACACCUGAAGCUAGCGCGUCUACCAAUUCCGCCACCUUCGCAUACCAUCAAUUCUUAAAAAGAAUUGCUACCACGGAGGCGCAUUCUA (((.(((.........((((....))))..........((.((....)).))...))).)))((..((((((........(((((((.....)))))))......)))))).))...... ( -29.84) >Sty_1 231 120 + 1 GGACUUGAACCCCCACGUCCGUAAGGACACUAACACCUGAAGCUAGCGCGUCUACCAAUUCCGCCACCUUCGCAUACCAUCGAUUCUUGAAAAGAAUCACUACCACGGAGGCGCAUUCUA (((.(((.........((((....))))..........((.((....)).))...))).)))((..((((((.........((((((.....)))))).......)))))).))...... ( -29.09) >consensus GGACUUGAACCCCCACGUCCGUAAGGACACUAACACCUGAAGCUAGCGCGUCUACCAAUUCCGCCACCUUCGCAUACCAUCAAUUCUUAAAAAGAAUUGCUACCACGGAGGCGCAUUCUA (((.(((.........((((....))))..........((.((....)).))...))).)))((..((((((........(((((((.....)))))))......)))))).))...... (-27.45 = -29.34 + 1.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:08:23 2006