
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,177,798 – 4,178,019 |
| Length | 221 |
| Max. P | 0.993284 |

| Location | 4,177,798 – 4,177,918 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -41.65 |
| Energy contribution | -41.10 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.80 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4177798 120 + 1 AAAGAUUUGUUCGUUGGAGCCUGGCCUAUCCAGGCCUCCGUCGAAGACCGCAGGAGUUUCGCAAGAAACUUAAUCCCCUGCGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUU ........((((...(((((((((.....))))).))))((((..(..((((((((((((....))))))......))))))....)..)))).))))...((((....))))....... ( -41.70) >Sfl_1 1 120 + 1 AAAGAUUUGUUCGUUGGAGCCUGGCCUAUCCAGGCCUCCGUCGAAGACCGCAGGAGUUUCGCAAGAAACUUAAUCCCCUGCGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUU ........((((...(((((((((.....))))).))))((((..(..((((((((((((....))))))......))))))....)..)))).))))...((((....))))....... ( -41.70) >Sty_1 1 119 + 1 AAGAAUUUGUUCGUUGGAGCCUGGCCUAUCCAGGCCUCCGUCGAAGACCGCAGGCGUCUCGCAAGAGGCUUAAUCCCCUGCGUAGACGGUGACAGAACGCUAAGAUUAUU-UUUUAUACU ........((((...(((((((((.....))))).))))((((..(..((((((.(((((....))))).......))))))....)..)))).))))............-......... ( -40.60) >consensus AAAGAUUUGUUCGUUGGAGCCUGGCCUAUCCAGGCCUCCGUCGAAGACCGCAGGAGUUUCGCAAGAAACUUAAUCCCCUGCGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUU ...(((((...(((((((((((((.....))))).))))((((..(..((((((((((((....))))))......))))))....)..))))..))))...)))))............. (-41.65 = -41.10 + -0.55)

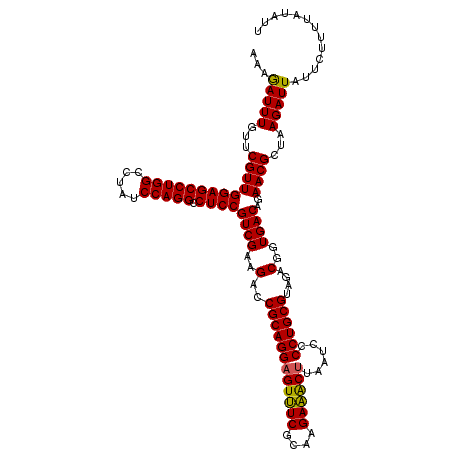
| Location | 4,177,798 – 4,177,918 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -37.24 |
| Energy contribution | -37.13 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4177798 120 - 1 AAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACGCAGGGGAUUAAGUUUCUUGCGAAACUCCUGCGGUCUUCGACGGAGGCCUGGAUAGGCCAGGCUCCAACGAACAAAUCUUU .......((((....))))...((((.....(((((..((((((((.((..((.....))..))))))))))......))))).(((((((.....))))))).....))))........ ( -38.50) >Sfl_1 1 120 - 1 AAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACGCAGGGGAUUAAGUUUCUUGCGAAACUCCUGCGGUCUUCGACGGAGGCCUGGAUAGGCCAGGCUCCAACGAACAAAUCUUU .......((((....))))...((((.....(((((..((((((((.((..((.....))..))))))))))......))))).(((((((.....))))))).....))))........ ( -38.50) >Sty_1 1 119 - 1 AGUAUAAAA-AAUAAUCUUAGCGUUCUGUCACCGUCUACGCAGGGGAUUAAGCCUCUUGCGAGACGCCUGCGGUCUUCGACGGAGGCCUGGAUAGGCCAGGCUCCAACGAACAAAUUCUU .........-............((((.((((((((((.((((((((.......))))))))))))(....))))....)))((((.(((((.....)))))))))...))))........ ( -41.30) >consensus AAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACGCAGGGGAUUAAGUUUCUUGCGAAACUCCUGCGGUCUUCGACGGAGGCCUGGAUAGGCCAGGCUCCAACGAACAAAUCUUU ......................((((.....(((((..((((((((.((..((.....))..))))))))))......))))).(((((((.....))))))).....))))........ (-37.24 = -37.13 + -0.11)


| Location | 4,177,838 – 4,177,958 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -39.93 |
| Consensus MFE | -38.46 |
| Energy contribution | -38.13 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4177838 120 + 1 UCGAAGACCGCAGGAGUUUCGCAAGAAACUUAAUCCCCUGCGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCC ..(((((.((((((((((((....))))))......))))))...((((((((((((.((((.((.(((......))).))))))....))))).)))))))...........))).)). ( -38.90) >Sfl_1 41 120 + 1 UCGAAGACCGCAGGAGUUUCGCAAGAAACUUAAUCCCCUGCGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCC ..(((((.((((((((((((....))))))......))))))...((((((((((((.((((.((.(((......))).))))))....))))).)))))))...........))).)). ( -38.90) >Sty_1 41 118 + 1 UCGAAGACCGCAGGCGUCUCGCAAGAGGCUUAAUCCCCUGCGUAGACGGUGACAGAACGCUAAGAUUAUU-UUUUAUACUCUGGCUUGUUUCUGCUCACCGUA-UUAAGACGCUCUUCUC ..(((((.((((((.(((((....))))).......))))))...((((((((((((.((((.((.(((.-....))).))))))....))))).))))))).-.........))))).. ( -42.00) >consensus UCGAAGACCGCAGGAGUUUCGCAAGAAACUUAAUCCCCUGCGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCC ........((((((((((((....))))))......))))))...((((((((((((.((((.((.(((......))).))))))....))))).))))))).................. (-38.46 = -38.13 + -0.33)

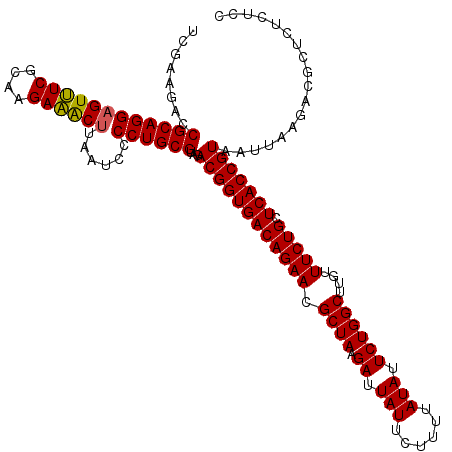

| Location | 4,177,838 – 4,177,958 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -40.73 |
| Consensus MFE | -37.06 |
| Energy contribution | -37.07 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4177838 120 - 1 GGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACGCAGGGGAUUAAGUUUCUUGCGAAACUCCUGCGGUCUUCGA (((((..((.........(((((((.(((((....((.(((.(((......))).)))..)).))))))))))))...((((((..((...))..))))))........))..))))).. ( -39.00) >Sfl_1 41 120 - 1 GGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACGCAGGGGAUUAAGUUUCUUGCGAAACUCCUGCGGUCUUCGA (((((..((.........(((((((.(((((....((.(((.(((......))).)))..)).))))))))))))...((((((..((...))..))))))........))..))))).. ( -39.00) >Sty_1 41 118 - 1 GAGAAGAGCGUCUUAA-UACGGUGAGCAGAAACAAGCCAGAGUAUAAAA-AAUAAUCUUAGCGUUCUGUCACCGUCUACGCAGGGGAUUAAGCCUCUUGCGAGACGCCUGCGGUCUUCGA ..(((((.(((.....-...(((((.(((((....((.(((.(((....-.))).)))..)).))))))))))((((.((((((((.......))))))))))))....))).))))).. ( -44.20) >consensus GGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACGCAGGGGAUUAAGUUUCUUGCGAAACUCCUGCGGUCUUCGA (((((..((.........(((((((.(((((....((.(((.(((......))).)))..)).))))))))))))...(((((((((.....)))))))))........))..))))).. (-37.06 = -37.07 + 0.01)



| Location | 4,177,878 – 4,177,997 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -36.87 |
| Energy contribution | -36.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4177878 119 + 1 CGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCCGUUUGGAGGAGUGAAGUGAGUUCCAGAG-AUUUUCUCUGG .....((((((((((((.((((.((.(((......))).))))))....))))).)))))))........(((((.((((....))))))))).........((((((-.....)))))) ( -42.30) >Sfl_1 81 119 + 1 CGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCCGUUUGGAGGAGUGAAGUGAGUUCCAGAG-AUUUUCUCUGG .....((((((((((((.((((.((.(((......))).))))))....))))).)))))))........(((((.((((....))))))))).........((((((-.....)))))) ( -42.30) >Sty_1 81 118 + 1 CGUAGACGGUGACAGAACGCUAAGAUUAUU-UUUUAUACUCUGGCUUGUUUCUGCUCACCGUA-UUAAGACGCUCUUCUCGUUGAGAUGAGUGAAGUGAGUUCCGGAACAUGAGUUCCGG .....((((((((((((.((((.((.(((.-....))).))))))....))))).))))))).-......(((((.((((...)))).))))).........(((((((....))))))) ( -40.80) >consensus CGUAGACGGUGACAGAACGCUAAGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCCGUUUGGAGGAGUGAAGUGAGUUCCAGAG_AUUUUCUCUGG .....((((((((((((.((((.((.(((......))).))))))....))))).)))))))........(((((.((((....))))))))).........((((((......)))))) (-36.87 = -36.77 + -0.11)

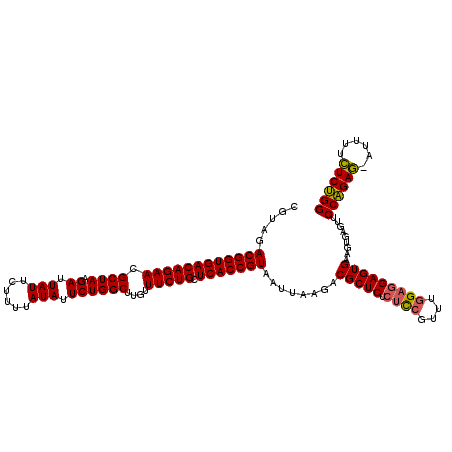

| Location | 4,177,878 – 4,177,997 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -32.96 |
| Energy contribution | -32.30 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4177878 119 - 1 CCAGAGAAAAU-CUCUGGAACUCACUUCACUCCUCCAAACGGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACG ((((((.....-)))))).........(.(((((((....)))).))).)........(((((((.(((((....((.(((.(((......))).)))..)).))))))))))))..... ( -37.30) >Sfl_1 81 119 - 1 CCAGAGAAAAU-CUCUGGAACUCACUUCACUCCUCCAAACGGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACG ((((((.....-)))))).........(.(((((((....)))).))).)........(((((((.(((((....((.(((.(((......))).)))..)).))))))))))))..... ( -37.30) >Sty_1 81 118 - 1 CCGGAACUCAUGUUCCGGAACUCACUUCACUCAUCUCAACGAGAAGAGCGUCUUAA-UACGGUGAGCAGAAACAAGCCAGAGUAUAAAA-AAUAAUCUUAGCGUUCUGUCACCGUCUACG (((((((....))))))).........(.(((.((((...)))).))).)......-.(((((((.(((((....((.(((.(((....-.))).)))..)).))))))))))))..... ( -36.40) >consensus CCAGAGAAAAU_CUCUGGAACUCACUUCACUCCUCCAAACGGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCUUAGCGUUCUGUCACCGUCUACG ((((((......)))))).........(.(((((((....)))).))).)........(((((((.(((((....((.(((.(((......))).)))..)).))))))))))))..... (-32.96 = -32.30 + -0.66)

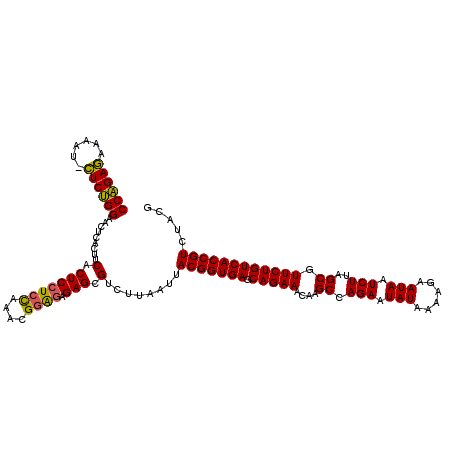
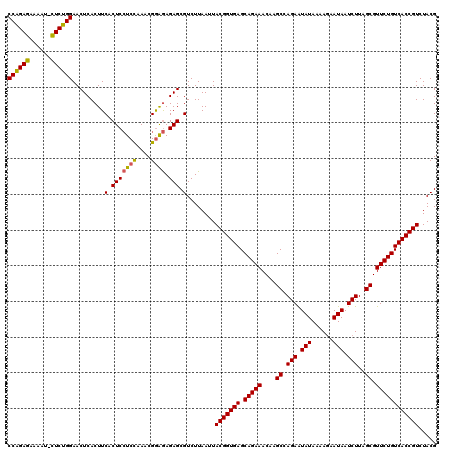
| Location | 4,177,900 – 4,178,019 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -35.07 |
| Energy contribution | -34.97 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4177900 119 + 1 AGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCCGUUUGGAGGAGUGAAGUGAGUUCCAGAG-AUUUUCUCUGGCAAACAUCCAGGAGCAAAGCUA (((.(((......))).)))((((((((((((((((((........).(((((.((((....)))))))))..)))))..((((((-.....))))))........))))))).))))). ( -40.50) >Sfl_1 103 119 + 1 AGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCCGUUUGGAGGAGUGAAGUGAGUUCCAGAG-AUUUUCUCUGGCAAACAUCCAGGAGCAAAGCUA (((.(((......))).)))((((((((((((((((((........).(((((.((((....)))))))))..)))))..((((((-.....))))))........))))))).))))). ( -40.50) >Sty_1 103 118 + 1 AGAUUAUU-UUUUAUACUCUGGCUUGUUUCUGCUCACCGUA-UUAAGACGCUCUUCUCGUUGAGAUGAGUGAAGUGAGUUCCGGAACAUGAGUUCCGGCAAACAUCCAGGAGCAAAGCUA (((.(((.-....))).)))((((((((((((((((((...-....).(((((.((((...)))).)))))..)))))..(((((((....)))))))........))))))).))))). ( -38.70) >consensus AGAUUAUUCUUUUAUAUUCUGGCUUGUUUCUGCUCACCGUAAUUAAGACGCUCUCUCCGUUUGGAGGAGUGAAGUGAGUUCCAGAG_AUUUUCUCUGGCAAACAUCCAGGAGCAAAGCUA (((.(((......))).)))((((((((((((((((((........).(((((.((((....)))))))))..)))))..((((((......))))))........))))))).))))). (-35.07 = -34.97 + -0.11)

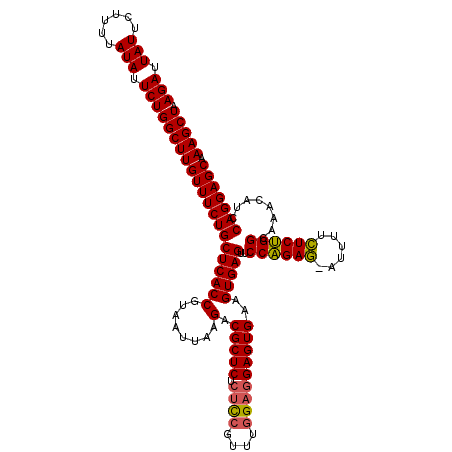

| Location | 4,177,900 – 4,178,019 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -29.16 |
| Energy contribution | -28.50 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4177900 119 - 1 UAGCUUUGCUCCUGGAUGUUUGCCAGAGAAAAU-CUCUGGAACUCACUUCACUCCUCCAAACGGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCU ...((((....((((.(((((.((((((.....-))))))..(((((....(((((((....)))).)))(((.......))))))))...)))))..))))......))))........ ( -34.20) >Sfl_1 103 119 - 1 UAGCUUUGCUCCUGGAUGUUUGCCAGAGAAAAU-CUCUGGAACUCACUUCACUCCUCCAAACGGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCU ...((((....((((.(((((.((((((.....-))))))..(((((....(((((((....)))).)))(((.......))))))))...)))))..))))......))))........ ( -34.20) >Sty_1 103 118 - 1 UAGCUUUGCUCCUGGAUGUUUGCCGGAACUCAUGUUCCGGAACUCACUUCACUCAUCUCAACGAGAAGAGCGUCUUAA-UACGGUGAGCAGAAACAAGCCAGAGUAUAAAA-AAUAAUCU ......(((((.(((.(((((.(((((((....)))))))..(((((....(((.((((...)))).)))(((.....-.))))))))...)))))..)))))))).....-........ ( -36.40) >consensus UAGCUUUGCUCCUGGAUGUUUGCCAGAGAAAAU_CUCUGGAACUCACUUCACUCCUCCAAACGGAGAGAGCGUCUUAAUUACGGUGAGCAGAAACAAGCCAGAAUAUAAAAGAAUAAUCU ...........((((.(((((.((((((......))))))..(((((....(((((((....)))).)))(((.......))))))))...)))))..)))).................. (-29.16 = -28.50 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:07:31 2006