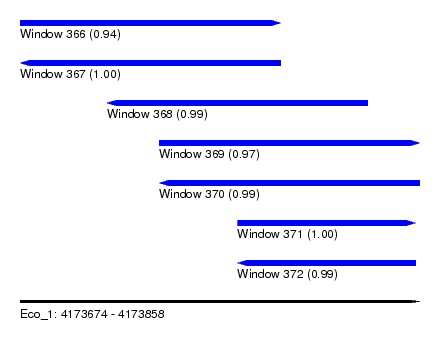
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,173,674 – 4,173,858 |
| Length | 184 |
| Max. P | 0.998607 |

| Location | 4,173,674 – 4,173,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -33.89 |
| Energy contribution | -33.00 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4173674 120 + 1 UCAGGUAGCCGAGUUCCAGGAUGCGGGCAUCGUAUAAUGGCUAUUACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUGCCCGCUCCAAGAUGUGCUGAUAUAGCUCAGUU ...((.(((...))))).(((.(((((((((((.....((......)).((((.......)))))))))(((((.......))))).))))))))).......(((((......))))). ( -39.90) >Hin_1 1 96 + 1 ---------------------UGCGGGCAUCGUAUAAUGGCUAUUACCUUAGCCUUCCAAGCUAAUGAUGCGGGUUCGAUUCCCGCUGCCCGCUCCAGA---CGCUGAUAUAGCUCAGUU ---------------------.(((((((.........(((((......)))))...............(((((.......))))))))))))......---.(((((......))))). ( -34.80) >Ype_1 1 120 + 1 UCAGGUAGCCGAGUUCUAAGAUGCGGGCAUCGUAUAAUGGCUAUUACCUCAGCCUUCCAAGCUGAUGAUGUGGGUUCGAUUCCCACUGCCCGCUCCAAGAUGUGCUGAUAUAGCUCAGUU ..............(((..((.(((((((((((.....((......)).((((.......)))))))))(((((.......))))).))))))))..)))...(((((......))))). ( -34.90) >consensus UCAGGUAGCCGAGUUC_A_GAUGCGGGCAUCGUAUAAUGGCUAUUACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUGCCCGCUCCAAGAUGUGCUGAUAUAGCUCAGUU ......................(((((((((((.....((......)).((((.......)))))))))(((((.......))))).))))))..........(((((......))))). (-33.89 = -33.00 + -0.89)



| Location | 4,173,674 – 4,173,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -42.40 |
| Consensus MFE | -38.82 |
| Energy contribution | -37.93 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4173674 120 - 1 AACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUAAUAGCCAUUAUACGAUGCCCGCAUCCUGGAACUCGGCUACCUGA ..(((((((.....)))...(((.(((((((((.(((((.......)))))(((.((((((.....))))))(((....))).......))))))))).))).)))..))))........ ( -46.80) >Hin_1 1 96 - 1 AACUGAGCUAUAUCAGCG---UCUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAUUAGCUUGGAAGGCUAAGGUAAUAGCCAUUAUACGAUGCCCGCA--------------------- ..((((......))))..---......((((((.(((((.......)))))(((.((((((.....))))))(((....))).......))))))))).--------------------- ( -36.90) >Ype_1 1 120 - 1 AACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGUGGGAAUCGAACCCACAUCAUCAGCUUGGAAGGCUGAGGUAAUAGCCAUUAUACGAUGCCCGCAUCUUAGAACUCGGCUACCUGA ..(((((((.....)))...(((.(((((((((.(((((.......)))))(((.((((((.....))))))(((....))).......))))))))).))).)))..))))........ ( -43.50) >consensus AACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUAAUAGCCAUUAUACGAUGCCCGCAUC_U_GAACUCGGCUACCUGA ..((((......))))...........((((((.(((((.......)))))(((.((((((.....))))))(((....))).......)))))))))...................... (-38.82 = -37.93 + -0.89)



| Location | 4,173,714 – 4,173,834 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -43.24 |
| Energy contribution | -43.47 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4173714 120 - 1 CGAACUGCCGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUAAUAG ....(((...((((((.((((.(((((.((.(((((((....((((......)))).......))))))).)).(((((.......)))))........))))))))).))))))..))) ( -47.80) >Hin_1 20 117 - 1 UGAACCGCCGACCUCACCCUUACCAAGGGUGCACUCUACCAACUGAGCUAUAUCAGCG---UCUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAUUAGCUUGGAAGGCUAAGGUAAUAG ...((((((.....((((((.....))))))(.(((((....((((......))))..---..))))).)))).(((((.......)))))....((((((.....)))))))))..... ( -40.60) >Ype_1 41 120 - 1 CGAACUGCCGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGUGGGAAUCGAACCCACAUCAUCAGCUUGGAAGGCUGAGGUAAUAG ....(((...((((((.((((.(((((.((.(((((((....((((......)))).......))))))).)).(((((.......)))))........))))))))).))))))..))) ( -46.30) >consensus CGAACUGCCGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUAAUAG ..........((((((.((((.(((((.((.(((((((....((((......)))).......))))))).)).(((((.......)))))........))))))))).))))))..... (-43.24 = -43.47 + 0.22)

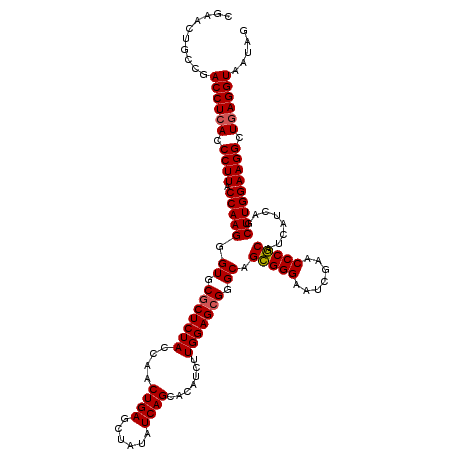

| Location | 4,173,738 – 4,173,858 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -46.53 |
| Consensus MFE | -44.25 |
| Energy contribution | -44.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4173738 120 + 1 AUGAUGCGGGUUCGAUUCCCGCUGCCCGCUCCAAGAUGUGCUGAUAUAGCUCAGUUGGUAGAGCGCACCCUUGGUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCUAUCAGCACCACUUCU .((..((((((.((.....))..))))))..))(((.(((((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))))....))) ( -49.10) >Hin_1 44 112 + 1 AUGAUGCGGGUUCGAUUCCCGCUGCCCGCUCCAGA---CGCUGAUAUAGCUCAGUUGGUAGAGUGCACCCUUGGUAAGGGUGAGGUCGGCGGUUCAAAUCCGCCUAUCAGCACCA----- .(((((((((((.((...(((((((((((((....---.(((((......))))).....)))))((((((.....)))))).)).)))))).)).)))))))..))))......----- ( -45.20) >Ype_1 65 120 + 1 AUGAUGUGGGUUCGAUUCCCACUGCCCGCUCCAAGAUGUGCUGAUAUAGCUCAGUUGGUAGAGCGCACCCUUGGUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCUAUCAGCACCAUUUCU .((..((((((............))))))..))(((((((((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))).))))).. ( -45.30) >consensus AUGAUGCGGGUUCGAUUCCCGCUGCCCGCUCCAAGAUGUGCUGAUAUAGCUCAGUUGGUAGAGCGCACCCUUGGUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCUAUCAGCACCA_UUCU .((..((((((............))))))..))....(((((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))))....... (-44.25 = -44.03 + -0.22)

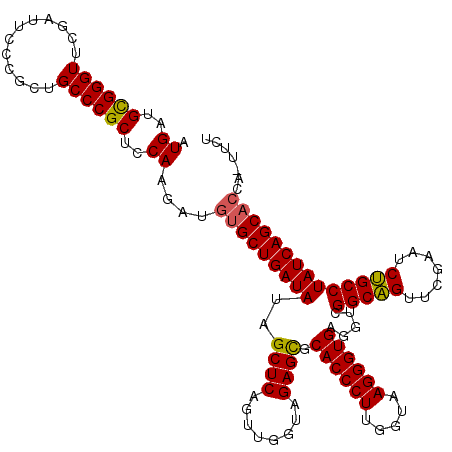

| Location | 4,173,738 – 4,173,858 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -45.77 |
| Consensus MFE | -42.45 |
| Energy contribution | -42.57 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4173738 120 - 1 AGAAGUGGUGCUGAUAGGCAGAUUCGAACUGCCGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAU ..(((..((((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))))..))).(((((((...((.....))..))))).)).. ( -48.60) >Hin_1 44 112 - 1 -----UGGUGCUGAUAGGCGGAUUUGAACCGCCGACCUCACCCUUACCAAGGGUGCACUCUACCAACUGAGCUAUAUCAGCG---UCUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAU -----.(..((((((((((((.......))))).....((((((.....))))))..(((........)))...))))))).---.)((..(((((...((.....))..)))))..)). ( -43.10) >Ype_1 65 120 - 1 AGAAAUGGUGCUGAUAGGCAGAUUCGAACUGCCGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGUGGGAAUCGAACCCACAUCAU .((..(((((((....))))(((((..((((((.....((((((.....))))))(((((((....((((......)))).......)))))))))))))..)))))....)))..)).. ( -45.60) >consensus AGAA_UGGUGCUGAUAGGCAGAUUCGAACUGCCGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCUAUAUCAGCACAUCUUGGAGCGGGCAGCGGGAAUCGAACCCGCAUCAU .......((((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))))....((..(((((...((.....))..)))))..)). (-42.45 = -42.57 + 0.12)



| Location | 4,173,774 – 4,173,856 |
|---|---|
| Length | 82 |
| Sequences | 4 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -35.55 |
| Energy contribution | -34.80 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.41 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4173774 82 + 1 UGUGCUGAUAUAGCUCAGUUGGUAGAGCGCACCCUUGGUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCUAUCAGCACCACUU .(((((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))))..... ( -35.40) >Sfl_1 1 81 + 1 -GUGCUGAUAUGGCUCAGUUGGUAGAGCGCACCCUUGGUAAGGGUGAGGUCCCCAGUUCGACUCUGGGUAUCAGCACCACUU -(((((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))))..... ( -34.60) >Sty_1 1 82 + 1 UGUGCUGAUAUGGCUCAGUUGGUAGAGCGCACCCUUGGUAAGGGUGAGGUCCCCAGUUCGACUCUGGGUAUCAGCACCACUU .(((((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))))..... ( -34.60) >Ype_1 1 79 + 1 UGUGCUGAUAUGGCUCAGUUGGUAGAGCGCACCCUUGGUAAGGGUGAGGUCCCCAGUUCGACUCUGGGUAUCAGCACCA--- .(((((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))))..--- ( -34.60) >consensus UGUGCUGAUAUGGCUCAGUUGGUAGAGCGCACCCUUGGUAAGGGUGAGGUCCCCAGUUCGACUCUGGGUAUCAGCACCACUU .(((((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))))..... (-35.55 = -34.80 + -0.75)

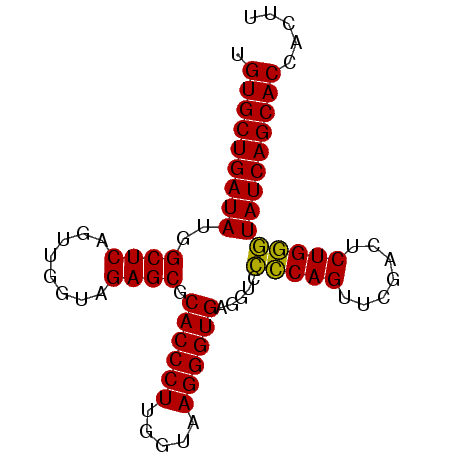

| Location | 4,173,774 – 4,173,856 |
|---|---|
| Length | 82 |
| Sequences | 4 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -35.65 |
| Energy contribution | -34.90 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.76 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4173774 82 - 1 AAGUGGUGCUGAUAGGCAGAUUCGAACUGCCGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCUAUAUCAGCACA .....((((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))). ( -36.00) >Sfl_1 1 81 - 1 AAGUGGUGCUGAUACCCAGAGUCGAACUGGGGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCCAUAUCAGCAC- .....((((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))))- ( -33.40) >Sty_1 1 82 - 1 AAGUGGUGCUGAUACCCAGAGUCGAACUGGGGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCCAUAUCAGCACA .....((((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))). ( -35.10) >Ype_1 1 79 - 1 ---UGGUGCUGAUACCCAGAGUCGAACUGGGGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCCAUAUCAGCACA ---..((((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))). ( -35.10) >consensus AAGUGGUGCUGAUACCCAGAGUCGAACUGGGGACCUCACCCUUACCAAGGGUGCGCUCUACCAACUGAGCCAUAUCAGCACA .....((((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))). (-35.65 = -34.90 + -0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:07:24 2006