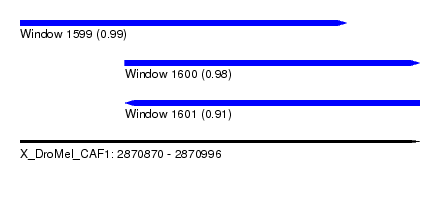
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,870,870 – 2,870,996 |
| Length | 126 |
| Max. P | 0.989029 |

| Location | 2,870,870 – 2,870,973 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -16.87 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
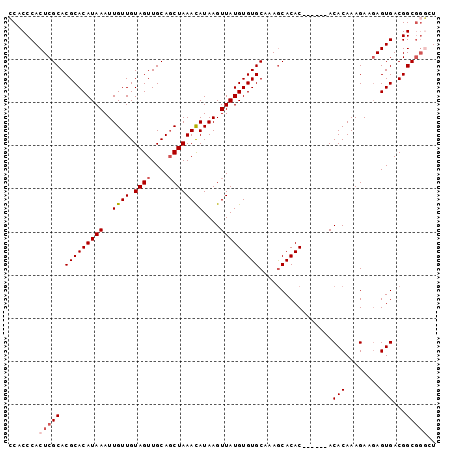
>X_DroMel_CAF1 2870870 103 + 22224390 ACCAUUUAACCCACUUUGGCCCCCUUAACCCAUCCACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAGCACAC------AC .............(((((((.......................)).(((((((((..((((.((((....))))))))....)))))))))))))).....------.. ( -17.50) >DroSec_CAF1 257806 109 + 1 CCCCUUUAACCCACUUUGGCCUCCCUAACCCAUCCACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAGCACACACACACAC .............(((((((.......................)).(((((((((..((((.((((....))))))))....))))))))))))))............. ( -17.50) >DroSim_CAF1 227776 109 + 1 CCCCUUUAACCCACUUUGGCCCUCUUAACCUAUCCACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAGCACACACACACAC .............(((((((.......................)).(((((((((..((((.((((....))))))))....))))))))))))))............. ( -17.50) >DroEre_CAF1 281005 98 + 1 CCCCUUUAACCCACUUUGGCCCCCUUA-A----ACACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAACACAC------AC ..........((.....))........-.----..........(((((((.......((((.((((....)))))))).......))))))).........------.. ( -15.54) >DroYak_CAF1 265845 99 + 1 CCCCUUUAACCCACUUUGGCCCCCUUAAA----ACACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAUCUAAAUAUAAGUUAUGUGUGCAAAGCACAC------AC .............(((((((.........----..........)).(((((((((.((((..(((......)))...)))).)))))))))))))).....------.. ( -16.31) >consensus CCCCUUUAACCCACUUUGGCCCCCUUAACC_AUCCACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAGCACAC______AC .............(((((((.......................)).(((((((((..((((.((((....))))))))....))))))))))))))............. (-15.56 = -15.80 + 0.24)

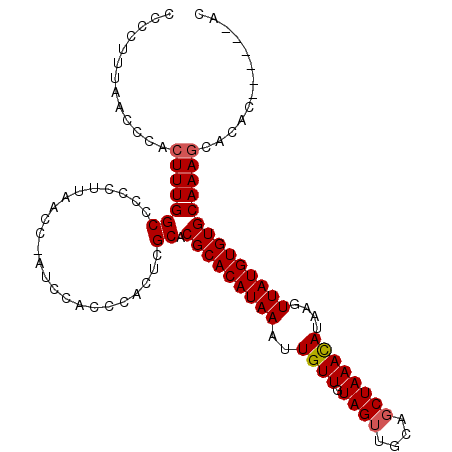
| Location | 2,870,903 – 2,870,996 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -20.28 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2870903 93 + 22224390 CCACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAGCACAC------ACACAAAGAACAGUGACGGCGGGCU .......(((((.(((((.......(((((....))))).............(((((((...))))))------)...........))).))))))).. ( -25.90) >DroSec_CAF1 257839 99 + 1 CCACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAGCACACACACACACACAAAGAAGAGUGACGGCGGGCU .......(((((.(((((.......(((((....))))).........((..(((((((...)))))))...))............))).))))))).. ( -26.00) >DroSim_CAF1 227809 99 + 1 CCACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAGCACACACACACACACAAAGAAGAGUGACGGCGGGCU .......(((((.(((((.......(((((....))))).........((..(((((((...)))))))...))............))).))))))).. ( -26.00) >DroEre_CAF1 281033 93 + 1 ACACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAACACAC------ACACAAAGAAGAGUGACGGCGGUCU .(.((((((((((((((.......((((.((((....)))))))).......))))))).........------.(.....)..)))))..)).).... ( -21.64) >DroYak_CAF1 265874 89 + 1 ACACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAUCUAAAUAUAAGUUAUGUGUGCAAAGCACAC------ACACAAAGAAGAGUGACGGC----U .....((((((((.(((((........))).)))))................(((((((...))))))------).........))))).....----. ( -21.10) >consensus CCACCCACUCGCACGCACAUAAAUUGUUGUAGUUGCAGCUAAACAUAAGUUAUGUGUGCAAAGCACAC______ACACAAAGAAGAGUGACGGCGGGCU .......(((((.(((((((((..((((.((((....))))))))....))))))))).................(((........)))...))))).. (-20.28 = -21.12 + 0.84)

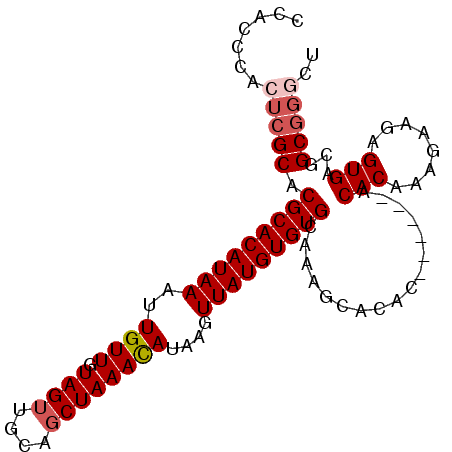
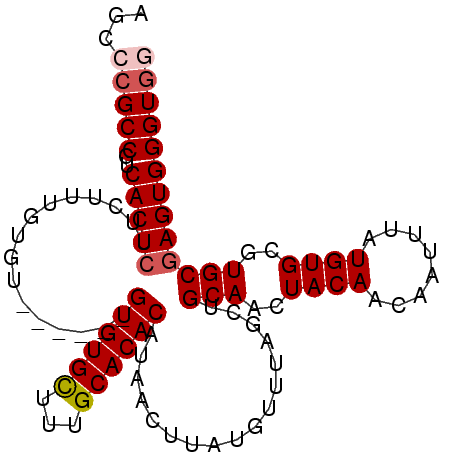
| Location | 2,870,903 – 2,870,996 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
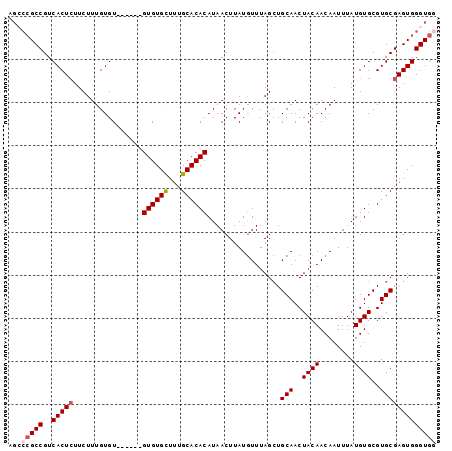
>X_DroMel_CAF1 2870903 93 - 22224390 AGCCCGCCGUCACUGUUCUUUGUGU------GUGUGCUUUGCACACAUAACUUAUGUUUAGCUGCAACUACAACAAUUUAUGUGCGUGCGAGUGGGUGG .((((((((.(((......((((((------((((.....))))))))))....(((((((......))).))))..........))))).)))))).. ( -30.00) >DroSec_CAF1 257839 99 - 1 AGCCCGCCGUCACUCUUCUUUGUGUGUGUGUGUGUGCUUUGCACACAUAACUUAUGUUUAGCUGCAACUACAACAAUUUAUGUGCGUGCGAGUGGGUGG ...(((((..(((((..((..(..((..(((((((((...)))))))))...))..)..))..(((..((((........))))..))))))))))))) ( -32.20) >DroSim_CAF1 227809 99 - 1 AGCCCGCCGUCACUCUUCUUUGUGUGUGUGUGUGUGCUUUGCACACAUAACUUAUGUUUAGCUGCAACUACAACAAUUUAUGUGCGUGCGAGUGGGUGG ...(((((..(((((..((..(..((..(((((((((...)))))))))...))..)..))..(((..((((........))))..))))))))))))) ( -32.20) >DroEre_CAF1 281033 93 - 1 AGACCGCCGUCACUCUUCUUUGUGU------GUGUGUUUUGCACACAUAACUUAUGUUUAGCUGCAACUACAACAAUUUAUGUGCGUGCGAGUGGGUGU ....((((..(((((....((((((------((((.....)))))))))).............(((..((((........))))..)))))))))))). ( -27.80) >DroYak_CAF1 265874 89 - 1 A----GCCGUCACUCUUCUUUGUGU------GUGUGCUUUGCACACAUAACUUAUAUUUAGAUGCAACUACAACAAUUUAUGUGCGUGCGAGUGGGUGU .----(((..(((((....((((((------((((.....)))))))))).............(((..((((........))))..))))))))))).. ( -26.00) >consensus AGCCCGCCGUCACUCUUCUUUGUGU______GUGUGCUUUGCACACAUAACUUAUGUUUAGCUGCAACUACAACAAUUUAUGUGCGUGCGAGUGGGUGG ...(((((..(((((................((((((...)))))).................(((..((((........))))..))))))))))))) (-22.98 = -23.62 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:47 2006