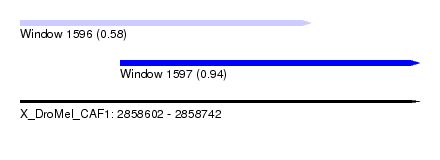
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,858,602 – 2,858,742 |
| Length | 140 |
| Max. P | 0.939882 |

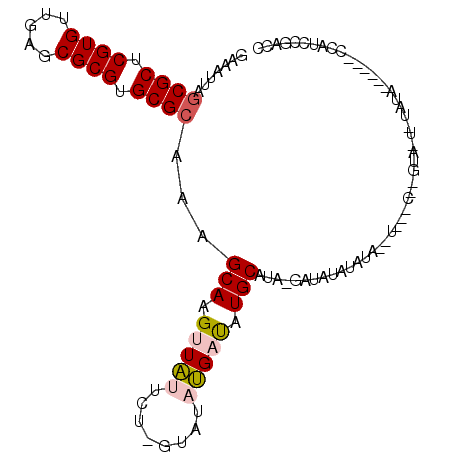
| Location | 2,858,602 – 2,858,704 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -17.70 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2858602 102 + 22224390 UGGUCUCGUUGCCUCUUGCAUUAAUUAAGAGCCCCGAAAUUAGCGCUCGUGUUAAGCGCGUGCGCAAAGCAAGUUAUUC--GUAUAUGAUAUGCAUA-CAUAUA----------U ((((.(((..((.(((((.......)))))))..))).))))((((.((((.....)))).))))...(((.(((((..--....))))).)))...-......----------. ( -27.20) >DroPse_CAF1 339609 92 + 1 ---------GG---CCCGCAUUAAUUAAGAGCC-AGAAAUUAGCGCUCGUGUUGCGCGCGUGCGCAGAGCAAGUUCAUCUUAUAUCGGUCUUGCAUCGGAUAUA----------U ---------((---(.(...........).)))-........((((.((((.....)))).)))).(((((((..(..........)..))))).)).......----------. ( -24.60) >DroSec_CAF1 246070 94 + 1 ---------GGCCUCUUGCAUUAAUUAAGAGCCCCGAAAUUAGCGCUCGUGUUGAGCGCGUGCGAAAAGCAAGUUAUUCU-GUAUAUGAUAUGCAUA-UAUAUA----------U ---------(((.(((((.......))))))))..(((....((((((.....)))))).(((.....))).....)))(-(((((((.....))))-))))..----------. ( -26.20) >DroSim_CAF1 215999 94 + 1 ---------GGCCUCUUGCAUUAAUUAAGAGCCCCGAAAUUAGCGCUCGUGUUGAGCGCGUGCGCAAAGCAAGUUAUUCC-GUAUAUGAUAUGCAUA-UAUAUA----------U ---------((...(((((..(((((..(.....)..)))))((((.((((.....)))).))))...))))).....))-(((((((.....))))-)))...----------. ( -26.60) >DroYak_CAF1 253835 101 + 1 ---------GGCCUC-UGCAUUAAUUAAGAGCCCCGAAAUUAGCGCUCGUGUUGAGCGCGUGCGCAAAGCAAGUUAU-CU-GCAUGUGAUAUGCAUA-UAUACUCGUACACAUA- ---------(((.((-(..........)))))).........((((.((((.....)))).))))...((.((((((-.(-(((((...)))))).)-)).))).)).......- ( -27.00) >DroPer_CAF1 359643 92 + 1 ---------GG---CCCGCAUUAAUUAAGAGCC-AGAAAUUAGCGCUCGUGUUGCGCGCGUGCGCAGAGCAAGUUCAUCUCAUAUCGGUCUUGCAUCGGAUAUA----------U ---------((---(.(...........).)))-........((((.((((.....)))).)))).(((((((..(..........)..))))).)).......----------. ( -24.60) >consensus _________GGCCUCUUGCAUUAAUUAAGAGCCCCGAAAUUAGCGCUCGUGUUGAGCGCGUGCGCAAAGCAAGUUAUUCU_GUAUAUGAUAUGCAUA_UAUAUA__________U ..........................................((((.((((.....)))).))))...(((.(((((........))))).)))..................... (-17.70 = -17.78 + 0.08)

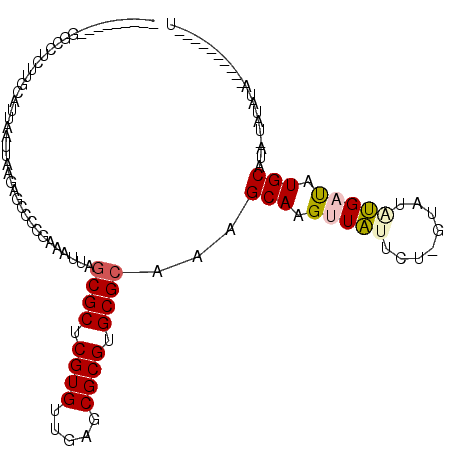
| Location | 2,858,637 – 2,858,742 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.47 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2858637 105 + 22224390 GAAAUUAGCGCUCGUGUUAAGCGCGUGCGCAAAGCAAGUUAUUC--GUAUAUGAUAUGCAUA-CAUAUAUAUACAUUCGCGAGUAAUAUAUACCCACCCGAUCCGACC .......((((.((((.....)))).)))).......(((((((--((..((((((((....-....))))).)))..)))))))))..................... ( -26.60) >DroPse_CAF1 339631 103 + 1 GAAAUUAGCGCUCGUGUUGCGCGCGUGCGCAGAGCAAGUUCAUCUUAUAUCGGUCUUGCAUCGGAUAUAUGUAUCUCAACUGGUAUUUUAUAU-----GCCGUUGGCA .......((((.((((.....)))).)))).(((((((..(..........)..))))).))(((((....)))))((((.(((((.....))-----)))))))... ( -31.00) >DroSec_CAF1 246096 83 + 1 GAAAUUAGCGCUCGUGUUGAGCGCGUGCGAAAAGCAAGUUAUUCU-GUAUAUGAUAUGCAUA-UAUAUAUAUA-----------------------UCCUAUCCGACC (((....((((((.....)))))).(((.....))).....)))(-(((((((.....))))-))))......-----------------------............ ( -19.40) >DroSim_CAF1 216025 83 + 1 GAAAUUAGCGCUCGUGUUGAGCGCGUGCGCAAAGCAAGUUAUUCC-GUAUAUGAUAUGCAUA-UAUAUAUAUA-----------------------UUCGAUCCGACC (((....((((.((((.....)))).))))...(((.(((((...-....))))).)))...-..........-----------------------)))......... ( -20.70) >DroPer_CAF1 359665 103 + 1 GAAAUUAGCGCUCGUGUUGCGCGCGUGCGCAGAGCAAGUUCAUCUCAUAUCGGUCUUGCAUCGGAUAUAUGUAUCUCAACUGGUAUUUUAUAU-----GCCGUUGGCA .......((((.((((.....)))).)))).(((((((..(..........)..))))).))(((((....)))))((((.(((((.....))-----)))))))... ( -31.00) >consensus GAAAUUAGCGCUCGUGUUGAGCGCGUGCGCAAAGCAAGUUAUUCU_GUAUAUGAUAUGCAUA_GAUAUAUAUA__U___C__GUA_U_UAUA______CCAUCCGACC .......((((.((((.....)))).))))...(((.(((((........))))).)))................................................. (-17.16 = -17.44 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:44 2006