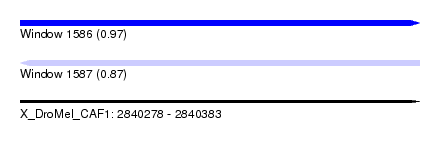
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,840,278 – 2,840,383 |
| Length | 105 |
| Max. P | 0.974454 |

| Location | 2,840,278 – 2,840,383 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.21 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2840278 105 + 22224390 UGUUUCUGCAGC--AGAAGGAGCAGCGCAACAGCAACACAUCUGCAGCACAGCUGCUCCUCAGAGUUCGUCAGAGGUCAGACUGCGAAAAAGAUGUGAAAAAAAAGA- ..((((.(((((--.((.((((((((......(((.......)))......)))))))))).((.(((....))).)).).))))))))..................- ( -32.00) >DroSec_CAF1 228202 98 + 1 UGUUUCUGCAGU--AGAAGCAGC-----AACAGCAACACAUCUGCAGCACAGCUGCUCCUCAGAGUUCGUCGGAGGUCAGACUGCUAAAAAGAUGUGAAAAAAAA--- ((((.((((...--....)))).-----))))....(((((((..((((...(((..((((.((.....)).)))).)))..))))....)))))))........--- ( -30.90) >DroSim_CAF1 201092 98 + 1 UGUUUCUGCAGC--AGAAGCAGC-----AUCAGCAACACAUCUGCAGCACAGCUGCUCCUCAGAGUUCGUCGGAGGUCAGACUGCGAAAAAGAUGUGAAAAAAAA--- ((((((((...)--)))))))((-----....))..(((((((....(.((((((..((((.((.....)).)))).))).))).)....)))))))........--- ( -32.80) >DroEre_CAF1 248477 92 + 1 UGUUUCUGCAGCAA----------------CAGCAAGACAUCUGCAGCACAGCUGCUCCACAGAGUUCGUCAGAGGUCAGACUGCGAAAAAGAUGUUGAAAAAAAAAA ..(((((((.....----------------..))).((((((((((((...))))).........((((.(((........)))))))..)))))))))))....... ( -22.40) >consensus UGUUUCUGCAGC__AGAAGCAGC_____AACAGCAACACAUCUGCAGCACAGCUGCUCCUCAGAGUUCGUCAGAGGUCAGACUGCGAAAAAGAUGUGAAAAAAAA___ (((....)))..........................(((((((....(.((((((..((((.((.....)).)))).))).))).)....)))))))........... (-21.45 = -21.57 + 0.12)

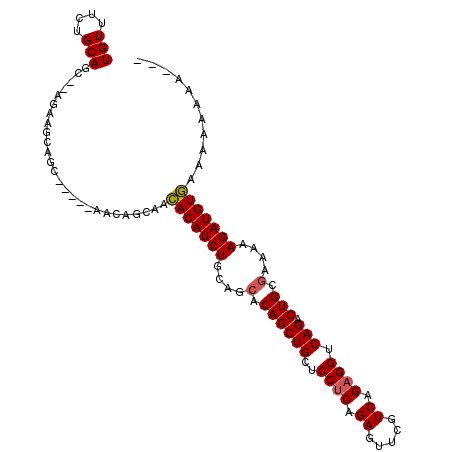
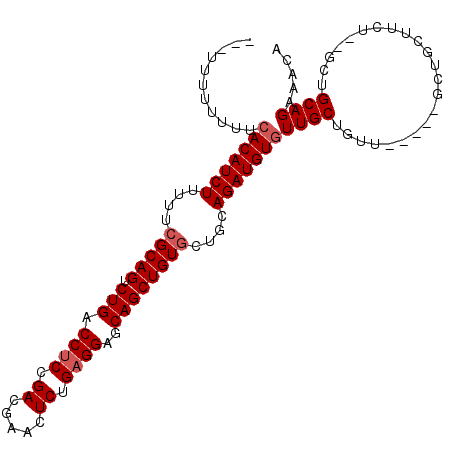
| Location | 2,840,278 – 2,840,383 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.21 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -22.30 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2840278 105 - 22224390 -UCUUUUUUUUCACAUCUUUUUCGCAGUCUGACCUCUGACGAACUCUGAGGAGCAGCUGUGCUGCAGAUGUGUUGCUGUUGCGCUGCUCCUUCU--GCUGCAGAAACA -..........(((((((.(..(((((.(((.((((.((.....)).))))..))))))))..).))))))).....(((...((((..(....--)..)))).))). ( -31.90) >DroSec_CAF1 228202 98 - 1 ---UUUUUUUUCACAUCUUUUUAGCAGUCUGACCUCCGACGAACUCUGAGGAGCAGCUGUGCUGCAGAUGUGUUGCUGUU-----GCUGCUUCU--ACUGCAGAAACA ---........(((((((.....((((.(((.((((.((.....)).))))..))))))).....))))))).....(((-----.((((....--...)))).))). ( -30.60) >DroSim_CAF1 201092 98 - 1 ---UUUUUUUUCACAUCUUUUUCGCAGUCUGACCUCCGACGAACUCUGAGGAGCAGCUGUGCUGCAGAUGUGUUGCUGAU-----GCUGCUUCU--GCUGCAGAAACA ---........(((((((.(..(((((.(((.((((.((.....)).))))..))))))))..).))))))).......(-----((.((....--)).)))...... ( -30.10) >DroEre_CAF1 248477 92 - 1 UUUUUUUUUUCAACAUCUUUUUCGCAGUCUGACCUCUGACGAACUCUGUGGAGCAGCUGUGCUGCAGAUGUCUUGCUG----------------UUGCUGCAGAAACA ....................(((((((........))).)))).((((..(((((((...((.......))...))))----------------)).)..)))).... ( -22.90) >consensus ___UUUUUUUUCACAUCUUUUUCGCAGUCUGACCUCCGACGAACUCUGAGGAGCAGCUGUGCUGCAGAUGUGUUGCUGUU_____GCUGCUUCU__GCUGCAGAAACA ...........(((((((....(((((.(((.((((.((.....)).))))..))))))))....)))))))((((.......................))))..... (-22.30 = -23.05 + 0.75)

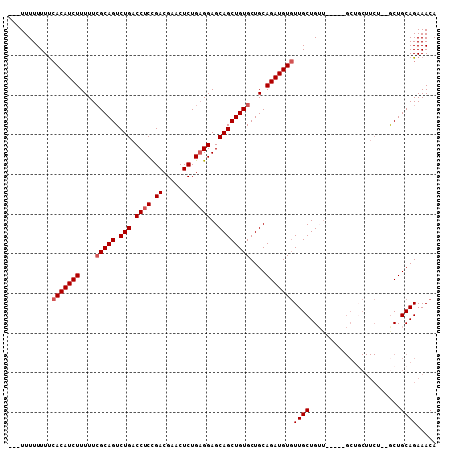
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:35 2006