| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,818,101 – 2,818,191 |
| Length | 90 |
| Max. P | 0.891865 |

| Location | 2,818,101 – 2,818,191 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -17.90 |
| Consensus MFE | -11.36 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2818101 90 + 22224390 AUAAUAUUUAGACAUAA---GUCCCAAAGAAACAAGGCAUACAUUUCUUCA----AUGCAAAAACAAGCUGCCUGAAAUGCAACGUAAUGCGUUGCA ..........(((....---)))...........(((((............----..((........)))))))....(((((((.....))))))) ( -15.93) >DroSec_CAF1 206491 81 + 1 AGAAUAU---------G---GCCCCAAAGAGACCAGGCAUACAUUUCUUCA----UUGCAAAAACAAGCUGCCUUAAAUGCAACGUAAUGCGUUGCA .......---------(---((....((((((...........))))))..----..((........)).))).....(((((((.....))))))) ( -14.80) >DroSim_CAF1 178404 90 + 1 AGAAUAUAUGGACAUAA---GCCCCAAAGAGACCAGGCAUACAUUUCCUGG----UUGCAAAAACAAGCUGCCUCAAAUGCAACGUAAUGCGUUGCA .........((.....(---((........(((((((.(.....).)))))----))..........))).)).....(((((((.....))))))) ( -21.97) >DroEre_CAF1 225900 90 + 1 AGAAUCU--ACACGAAAAAAAUGCC-AACAAAUCAGGCAUACAUUUCUUCA----GUGCCGAAGAAAGCUGGCUGAAAUGCAAUGUAAUGCGUUGCA .......--................-......((((.((....(((((((.----.....)))))))..)).))))..(((((((.....))))))) ( -21.50) >DroYak_CAF1 213087 91 + 1 GGAAUUU--ACACGAAA---ACCCC-AACAAACCAGGCUUACAUUUUUUAAAAGAAAGCCAAAGAAAGCUGCCUAAAAUGCAACGUAAUGCGUUGCA ((..(((--......))---)..))-.........(((((...(((....)))..)))))..................(((((((.....))))))) ( -15.30) >consensus AGAAUAU__ACACAAAA___GCCCCAAAGAAACCAGGCAUACAUUUCUUCA____AUGCAAAAACAAGCUGCCUGAAAUGCAACGUAAUGCGUUGCA ..................................(((((..................((........)))))))....(((((((.....))))))) (-11.36 = -11.60 + 0.24)

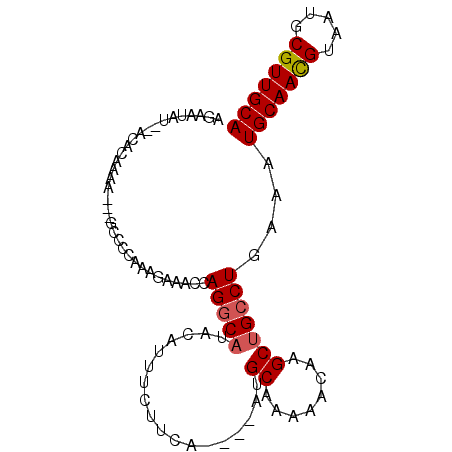
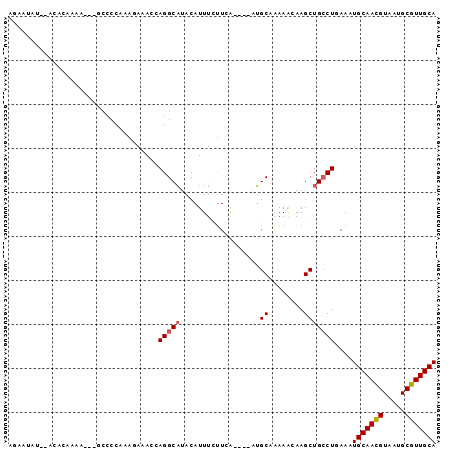
| Location | 2,818,101 – 2,818,191 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -12.12 |
| Energy contribution | -13.20 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2818101 90 - 22224390 UGCAACGCAUUACGUUGCAUUUCAGGCAGCUUGUUUUUGCAU----UGAAGAAAUGUAUGCCUUGUUUCUUUGGGAC---UUAUGUCUAAAUAUUAU (((((((.....)))))))....(((((((........)).(----..((((((((.......))))))))..)...---...)))))......... ( -22.10) >DroSec_CAF1 206491 81 - 1 UGCAACGCAUUACGUUGCAUUUAAGGCAGCUUGUUUUUGCAA----UGAAGAAAUGUAUGCCUGGUCUCUUUGGGGC---C---------AUAUUCU (((((((.....)))))))....(((((...(((((((....----...)))))))..)))))(((((.....))))---)---------....... ( -25.70) >DroSim_CAF1 178404 90 - 1 UGCAACGCAUUACGUUGCAUUUGAGGCAGCUUGUUUUUGCAA----CCAGGAAAUGUAUGCCUGGUCUCUUUGGGGC---UUAUGUCCAUAUAUUCU (((((((.....)))))))...(((((.((........))..----.((((.........)))))))))....((((---....))))......... ( -26.60) >DroEre_CAF1 225900 90 - 1 UGCAACGCAUUACAUUGCAUUUCAGCCAGCUUUCUUCGGCAC----UGAAGAAAUGUAUGCCUGAUUUGUU-GGCAUUUUUUUCGUGU--AGAUUCU ((((..(((......)))......((((((..((...((((.----...(....)...)))).))...)))-)))..........)))--)...... ( -20.90) >DroYak_CAF1 213087 91 - 1 UGCAACGCAUUACGUUGCAUUUUAGGCAGCUUUCUUUGGCUUUCUUUUAAAAAAUGUAAGCCUGGUUUGUU-GGGGU---UUUCGUGU--AAAUUCC (((((((.....)))))))....(((.((((......)))).)))((((....(((.((((((.(.....)-.))))---)).))).)--))).... ( -21.30) >consensus UGCAACGCAUUACGUUGCAUUUCAGGCAGCUUGUUUUUGCAA____UGAAGAAAUGUAUGCCUGGUUUCUUUGGGGC___UUACGUCU__AUAUUCU (((((((.....)))))))....(((((...((((((.............))))))..))))).................................. (-12.12 = -13.20 + 1.08)


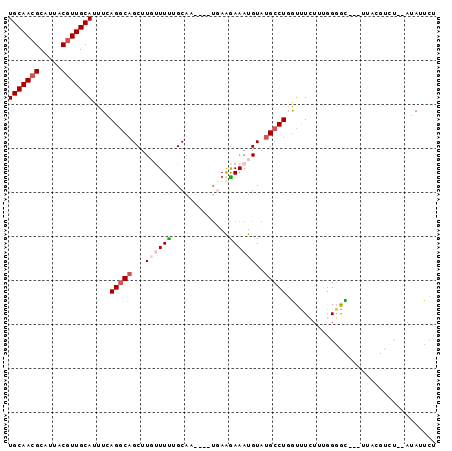
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:29 2006