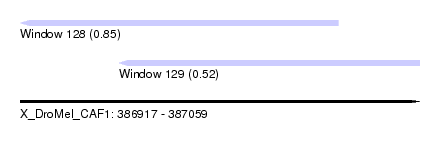
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 386,917 – 387,059 |
| Length | 142 |
| Max. P | 0.850230 |

| Location | 386,917 – 387,030 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -23.85 |
| Energy contribution | -23.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
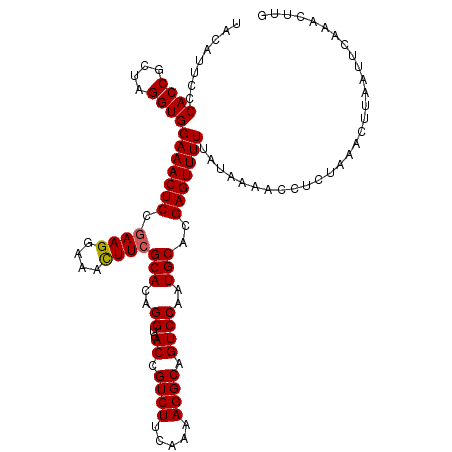
>X_DroMel_CAF1 386917 113 - 22224390 UACAUGCACACCGCUAGGUGGAAACUCCCAAGGGAAUUUCGCACAGGUCACCGUCUUCAAAAGGCUGUCCUAUGCACGAGUUUUUAUAA-ACCACUAAACUUAAUUCAAAUUUG ........((((....))))((((.(((....))).))))(((.(((.((..((((.....)))))).))).)))..((((((......-......))))))............ ( -26.70) >DroSec_CAF1 20780 114 - 1 UACAUUCCCACCGCUAGGUGGAAACUCCGAAGGAAACUUCGCACAGGUCACCGUCUUCAAAAGGCAGUCCAAUGCACGAGUUUUUAUAAAACCUCUAAACUUAAUUCAAACUUG .......(((((....)))))((((((.((((....))))(((..((..((.((((.....)))).))))..)))..))))))............................... ( -29.40) >DroSim_CAF1 21026 114 - 1 UACAUUCCCACCGCUAGGUGGAAACUCCGAAGGAAACUUCGCACAGGCCACUGUCUUCAAAAGGCAGUCCAAUGCACGAGUUUUUAUAAAACCUCUAAACUUAAUUCAAACUUG .......(((((....)))))((((((.((((....))))(((..((..(((((((.....)))))))))..)))..))))))............................... ( -34.00) >consensus UACAUUCCCACCGCUAGGUGGAAACUCCGAAGGAAACUUCGCACAGGUCACCGUCUUCAAAAGGCAGUCCAAUGCACGAGUUUUUAUAAAACCUCUAAACUUAAUUCAAACUUG ........((((....))))(((((((.((((....))))(((..((..((.((((.....)))).))))..)))..))))))).............................. (-23.85 = -23.97 + 0.11)

| Location | 386,952 – 387,059 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.29 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -28.61 |
| Energy contribution | -28.07 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
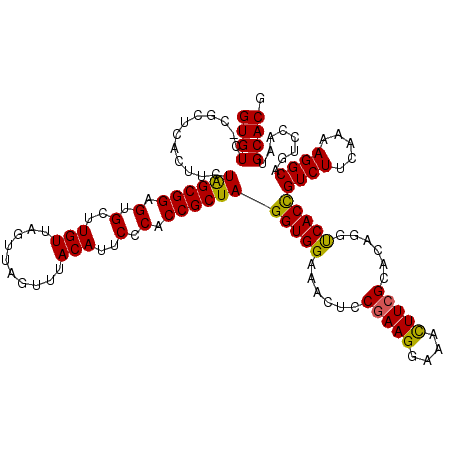
>X_DroMel_CAF1 386952 107 - 22224390 GUGUC-----------UGGCGGAGUGCUUGUUAGUUAGUUUACAUGCACACCGCUAGGUGGAAACUCCCAAGGGAAUUUCGCACAGGUCACCGUCUUCAAAAGGCUGUCCUAUGCACG (((((-----------((((((.((((.(((..........))).)))).)))))))(((((((.(((....))).)))).)))(((.((..((((.....)))))).)))..)))). ( -41.00) >DroSec_CAF1 20816 118 - 1 GUGUCGCGCUCACUUCUAGCGGAGUGCUUGUUAGUUAGUUUACAUUCCCACCGCUAGGUGGAAACUCCGAAGGAAACUUCGCACAGGUCACCGUCUUCAAAAGGCAGUCCAAUGCACG ((((...(((.......)))(((((((((((................(((((....)))))......(((((....))))).))))).))).((((.....))))..)))...)))). ( -36.10) >DroSim_CAF1 21062 118 - 1 GUGUCUCGCUCACUUCUAGCGGAGUGUUUGUUAGUUAGUUUACAUUCCCACCGCUAGGUGGAAACUCCGAAGGAAACUUCGCACAGGCCACUGUCUUCAAAAGGCAGUCCAAUGCACG ((((..............(((((((........((......))....(((((....)))))..)))))((((....))))))...((..(((((((.....)))))))))...)))). ( -39.00) >consensus GUGUC_CGCUCACUUCUAGCGGAGUGCUUGUUAGUUAGUUUACAUUCCCACCGCUAGGUGGAAACUCCGAAGGAAACUUCGCACAGGUCACCGUCUUCAAAAGGCAGUCCAAUGCACG ((((............((((((.(.(..(((..........)))..).).))))))(((((......(((((....)))))......)))))((((.....))))........)))). (-28.61 = -28.07 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:13 2006