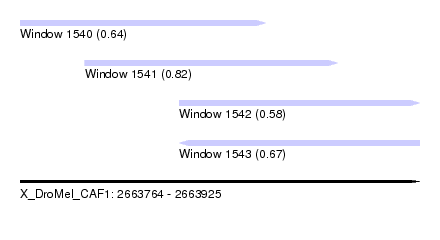
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,663,764 – 2,663,925 |
| Length | 161 |
| Max. P | 0.822858 |

| Location | 2,663,764 – 2,663,863 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -16.73 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2663764 99 + 22224390 -C----ACGCCGCC--------CCCCG-CCCGCUAAGCCCAAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCAGAAGCGCU-UUGUUUCUGUGCAACAAACAAGU -.----..((.((.--------....)-)..))...................((((((((...((((.....))))...((((((((...-..)))))))).....)))))))) ( -17.20) >DroGri_CAF1 103370 106 + 1 GUAGCUCUGCAUCU--------CGGCUUGAGCCUAAACCCAAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCGGAAGCGCUUUUGUUUCUGUGCAACAAACAAGU ...((((.((....--------..))..))))....................((((((((.((.(((...(((...((((((....)))))))))....)))))..)))))))) ( -24.10) >DroSec_CAF1 70099 107 + 1 -C----CCACCACCCCAACCCGCCCCG-CCCGCUAAGCCCAAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCAGAAGCGCU-UUGUUUCUGUGCAACAAACAAGU -.----.....................-........................((((((((...((((.....))))...((((((((...-..)))))))).....)))))))) ( -16.10) >DroEre_CAF1 80855 91 + 1 -U--------------------CCAUA-GCCGCUAAGCCCGAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCAGAAGCGCU-UUGUUUCUGUGCAACAAACAAGU -.--------------------.((((-(..((.((((..(....)..........((((((((..(((.....)))..)))))))))))-).))..)))))............ ( -17.50) >DroYak_CAF1 76767 91 + 1 -U--------------------UCAUU-CCCGCUAAGCCCAAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCAGAAGCGCU-UUGUUUCUGUGCAACAAACAAGU -.--------------------.....-........................((((((((...((((.....))))...((((((((...-..)))))))).....)))))))) ( -16.10) >DroMoj_CAF1 128355 101 + 1 -UA--GCUA--UCU--------CUGUUUAAGCCUAAGCUCAAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCGGAAACGCUUUUGUUUCUGUGCAACAAACAAGU -.(--(((.--...--------((.....))....)))).............((((((((.((.(((...(((...((((((....)))))))))....)))))..)))))))) ( -21.80) >consensus _U____C_____C_________CCACU_CCCGCUAAGCCCAAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCAGAAGCGCU_UUGUUUCUGUGCAACAAACAAGU ....................................................((((((((...((((.....))))...(((((((((....))))))))).....)))))))) (-16.73 = -16.37 + -0.36)

| Location | 2,663,790 – 2,663,892 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.12 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2663790 102 + 22224390 AAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCAGAAGCGCU-UUGUUUCUGUGCAACAAACAAGUUUCCCUGCC-----------UCGCUGCCAACUGGCACCUC ............((((((((...((((.....))))...((((((((...-..)))))))).....)))))))).........-----------..(.((((....)))).).. ( -21.60) >DroVir_CAF1 106084 110 + 1 AAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCGGAAGCGCUUUUGUUUCUGUGCAACAAACAAGUUUCUUUGGGCCCAGAGUCAGUCGCUGCCA-CUGCCAC--- ....(((.....((((((((.((.(((...(((...((((((....)))))))))....)))))..)))))))).....)))...(((.(.(((...))).).-)))....--- ( -23.80) >DroPse_CAF1 109213 83 + 1 AAAACCAUUAACACUUGUUUCUGUCAUUGUACAUGAAAAGCGGAAGCGCUUUUGUUUCUGUGCAACAAACAAGUUUCCUUUCG------------------------------- ............(((((((..(((...((((((.((((((((....))))....)))))))))))))))))))).........------------------------------- ( -19.60) >DroGri_CAF1 103402 108 + 1 AAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCGGAAGCGCUUUUGUUUCUGUGCAACAAACAAGUUUUCUUGAGCCCA--GUUCGUCGACGACG-CCGCCCC--- .....((..((.((((((((.((.(((...(((...((((((....)))))))))....)))))..)))))))).))..)).((...--((.((....)))).-..))...--- ( -21.50) >DroMoj_CAF1 128382 108 + 1 AAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCGGAAACGCUUUUGUUUCUGUGCAACAAACAAGUUUCUUUGCGCCCA--GUCAGUGGCUGCCA-CUGCCAC--- ............((((((((.((.(((...(((...((((((....)))))))))....)))))..)))))))).............--(.((((((...)))-))).)..--- ( -27.20) >DroPer_CAF1 126733 83 + 1 AAAACCAUUAACACUUGUUUCUGUCAUUGUACAUGAAAAGCGGAAGCGCUUUUGUUUCUGUGCAACAAACAAGUUUCCUUUCG------------------------------- ............(((((((..(((...((((((.((((((((....))))....)))))))))))))))))))).........------------------------------- ( -19.60) >consensus AAAACCAUUAACACUUGUUUCUGUCAUUUUACAUGAAAAGCGGAAGCGCUUUUGUUUCUGUGCAACAAACAAGUUUCCUUGCG___________UCGCUGCCA_CUGCCAC___ ............((((((((...((((.....))))...(((((((((....))))))))).....))))))))........................................ (-16.39 = -16.12 + -0.28)
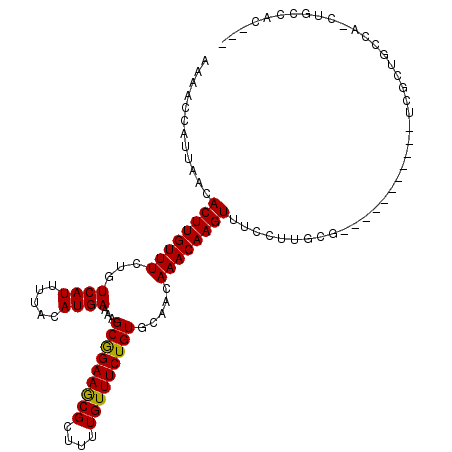
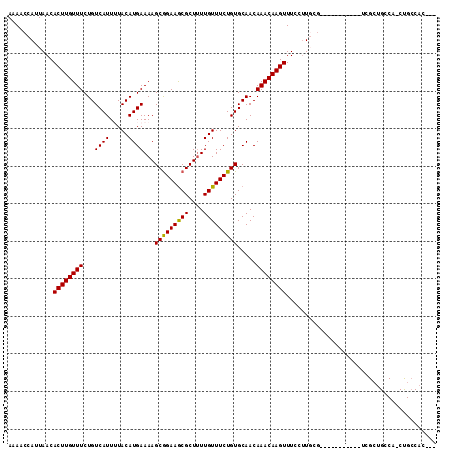
| Location | 2,663,828 – 2,663,925 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -22.30 |
| Energy contribution | -23.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2663828 97 + 22224390 AGCAGAAGCGCUUUGUUUCUGUGCAACAAACAAGUUUCCCUGCCUCGCUGCCAACUGGCACCUCGCAACUGGCAACCGGCA-------ACCGGCAACUGGCAAC .((((((((.....))))))))((........((((.((.((((..(.(((((....((.....))...))))).).))))-------...)).)))).))... ( -28.00) >DroSec_CAF1 70171 97 + 1 AGCAGAAGCGCUUUGUUUCUGUGCAACAAACAAGUUUCCUUGCCACGCUGCCAACUGGCACCUCGCAACCGGCAACCGGCA-------ACCGGCAACUGGCAAC .((((((((.....)))))))).................((((((.(.((((.....((.....))..((((...))))..-------...)))).))))))). ( -29.60) >DroEre_CAF1 80911 104 + 1 AGCAGAAGCGCUUUGUUUCUGUGCAACAAACAAGUUUCCUUGCCUCGCCGCCAACUGGCACCUCGCAACUGGCAACCGGCAACUGGCAACUGGCAACUGGCAAC .((((((((.....))))))))((........((((.(((((((..((((((....))).....((.....))....)))....)))))..)).)))).))... ( -32.00) >DroYak_CAF1 76823 90 + 1 AGCAGAAGCGCUUUGUUUCUGUGCAACAAACAAGUUUCCUUGCCUCGCUGCCAACUGGCACCUCGCAACUG--------------GUAACUGGUAACUGGCAAC .((((((((.....))))))))((.....((.((((.((((((...(.((((....)))).)..))))..)--------------).)))).)).....))... ( -25.30) >consensus AGCAGAAGCGCUUUGUUUCUGUGCAACAAACAAGUUUCCUUGCCUCGCUGCCAACUGGCACCUCGCAACUGGCAACCGGCA_______ACCGGCAACUGGCAAC .((((((((.....))))))))...........(((.(((((((..((((((....))))....))..((((...))))............)))))..)).))) (-22.30 = -23.30 + 1.00)

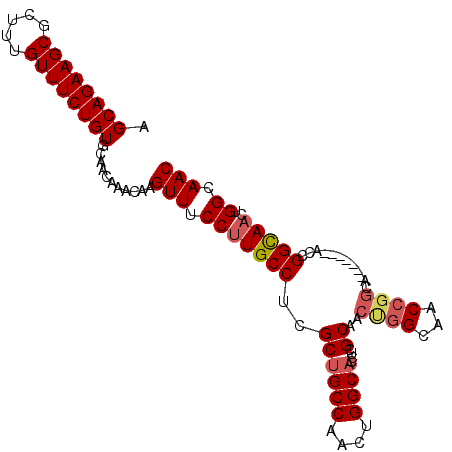
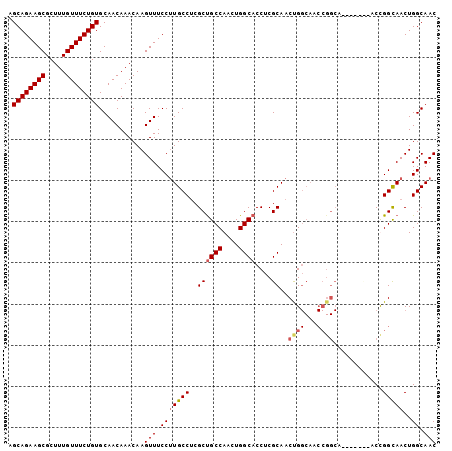
| Location | 2,663,828 – 2,663,925 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -31.77 |
| Energy contribution | -33.90 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2663828 97 - 22224390 GUUGCCAGUUGCCGGU-------UGCCGGUUGCCAGUUGCGAGGUGCCAGUUGGCAGCGAGGCAGGGAAACUUGUUUGUUGCACAGAAACAAAGCGCUUCUGCU ((.((..((.(((((.-------..))))).))..)).))(((((((..(((.((((((((.((((....)))))))))))).....)))...))))))).... ( -37.90) >DroSec_CAF1 70171 97 - 1 GUUGCCAGUUGCCGGU-------UGCCGGUUGCCGGUUGCGAGGUGCCAGUUGGCAGCGUGGCAAGGAAACUUGUUUGUUGCACAGAAACAAAGCGCUUCUGCU ((.(((.((.(((((.-------..))))).)).))).))(((((((..(((.((((((.((((((....)))))))))))).....)))...))))))).... ( -41.30) >DroEre_CAF1 80911 104 - 1 GUUGCCAGUUGCCAGUUGCCAGUUGCCGGUUGCCAGUUGCGAGGUGCCAGUUGGCGGCGAGGCAAGGAAACUUGUUUGUUGCACAGAAACAAAGCGCUUCUGCU ((.((..((.(((.((........)).))).))..)).))(((((((..(((.((((((((.((((....)))))))))))).....)))...))))))).... ( -34.70) >DroYak_CAF1 76823 90 - 1 GUUGCCAGUUACCAGUUAC--------------CAGUUGCGAGGUGCCAGUUGGCAGCGAGGCAAGGAAACUUGUUUGUUGCACAGAAACAAAGCGCUUCUGCU ...................--------------.....(((((((((..(((.((((((((.((((....)))))))))))).....)))...))))))).)). ( -29.40) >consensus GUUGCCAGUUGCCAGU_______UGCCGGUUGCCAGUUGCGAGGUGCCAGUUGGCAGCGAGGCAAGGAAACUUGUUUGUUGCACAGAAACAAAGCGCUUCUGCU ((.((..((.((((((........)))))).))..)).))(((((((..(((.((((((((.((((....)))))))))))).....)))...))))))).... (-31.77 = -33.90 + 2.13)

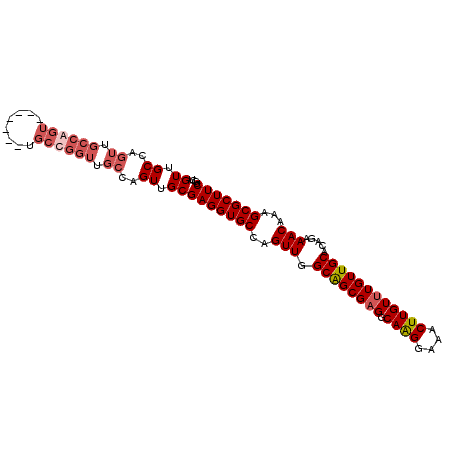
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:54 2006