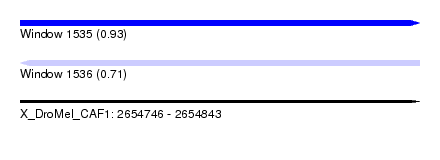
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,654,746 – 2,654,843 |
| Length | 97 |
| Max. P | 0.934511 |

| Location | 2,654,746 – 2,654,843 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2654746 97 + 22224390 AAGUAAAUCAGCGCUGCCAUUUGCCAUGCCCCGAAAAGUACGAUCCAUGG-AAAAAGUGGCAAAAG-UGGUAAAUGGUCGGUGGUGGUGGUGGGGGGGU ......((((.(((((((((((((((((((.((.......)).((....)-)......))))....-)))))))))).))))).))))........... ( -30.40) >DroPse_CAF1 97125 78 + 1 AAGUAAAUCAGCGCUGCCAUUUGCCAUGCC-CGAAAAGUAUGCUCCAUGG-AUAAAGUGGCAGUUAACA--G-------------GG----UGGCAGGG ...........(.((((((((((((((..(-((...((....))...)))-.....)))))))......--.-------------.)----)))))).) ( -20.20) >DroSec_CAF1 61400 75 + 1 AAGUAAAUCAGCGCUGCCAUUUGCCAUGGC-CAAAAAGUACGAUCCAUGGGAAAAAGUGGCAAAUG-UG----------------------CGGCAGGG ............(((((((((((((((..(-(.................)).....))))))))))-.)----------------------)))).... ( -24.53) >DroEre_CAF1 71197 80 + 1 AAGUAAAUCAGCGCUGCCAUUUGCCAUGCC-CAAAAAGUACGAUCCAUGG-AAAAAGUGGCAAAUG-UGGAGG--------GA--------GGGCAGGA .......((..(.((.(((((((((((..(-((..............)))-.....))))))))))-.).)).--------).--------.))..... ( -19.64) >DroYak_CAF1 67397 75 + 1 AAGUAAAUCAGCGCUGCCAUUUGCCAUGCC-CAAAAAAAAGGAUCCAUGG-AAAAAGUGGCAAAUG-UGCGG---------AA--------G----CGG ..........((.((((((((((((((..(-(........)).((....)-)....))))))))))-.))))---------..--------)----).. ( -24.10) >DroPer_CAF1 114662 78 + 1 AAGUAAAUCAGCGCUGCCAUUUGCCAUGCC-CGAAAAGUAUGCUCCAUGG-AUAAAGUGGCAGUUAACA--G-------------GG----UGGCAGGG ...........(.((((((((((((((..(-((...((....))...)))-.....)))))))......--.-------------.)----)))))).) ( -20.20) >consensus AAGUAAAUCAGCGCUGCCAUUUGCCAUGCC_CAAAAAGUACGAUCCAUGG_AAAAAGUGGCAAAUG_UG__G___________________GGGCAGGG ..............((((((((.(((((.................)))))....))))))))..................................... (-13.90 = -13.90 + 0.00)



| Location | 2,654,746 – 2,654,843 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -18.12 |
| Consensus MFE | -11.53 |
| Energy contribution | -11.31 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2654746 97 - 22224390 ACCCCCCCACCACCACCACCGACCAUUUACCA-CUUUUGCCACUUUUU-CCAUGGAUCGUACUUUUCGGGGCAUGGCAAAUGGCAGCGCUGAUUUACUU ................((.((.((((((.(((-....((((......(-(....))(((.......)))))))))).))))))...)).))........ ( -17.20) >DroPse_CAF1 97125 78 - 1 CCCUGCCA----CC-------------C--UGUUAACUGCCACUUUAU-CCAUGGAGCAUACUUUUCG-GGCAUGGCAAAUGGCAGCGCUGAUUUACUU ..((((((----..-------------.--(((((..((((.(((((.-...)))))....(.....)-)))))))))..))))))............. ( -16.30) >DroSec_CAF1 61400 75 - 1 CCCUGCCG----------------------CA-CAUUUGCCACUUUUUCCCAUGGAUCGUACUUUUUG-GCCAUGGCAAAUGGCAGCGCUGAUUUACUU ....((.(----------------------(.-(((((((((..........(((.(((.......))-))))))))))))))).))............ ( -19.80) >DroEre_CAF1 71197 80 - 1 UCCUGCCC--------UC--------CCUCCA-CAUUUGCCACUUUUU-CCAUGGAUCGUACUUUUUG-GGCAUGGCAAAUGGCAGCGCUGAUUUACUU ..(((((.--------..--------......-.((((((((.....(-(((..((......))..))-))..)))))))))))))............. ( -17.30) >DroYak_CAF1 67397 75 - 1 CCG----C--------UU---------CCGCA-CAUUUGCCACUUUUU-CCAUGGAUCCUUUUUUUUG-GGCAUGGCAAAUGGCAGCGCUGAUUUACUU .((----(--------(.---------..((.-(((((((((.....(-(((..((......))..))-))..)))))))))))))))........... ( -21.80) >DroPer_CAF1 114662 78 - 1 CCCUGCCA----CC-------------C--UGUUAACUGCCACUUUAU-CCAUGGAGCAUACUUUUCG-GGCAUGGCAAAUGGCAGCGCUGAUUUACUU ..((((((----..-------------.--(((((..((((.(((((.-...)))))....(.....)-)))))))))..))))))............. ( -16.30) >consensus CCCUGCCC___________________C__CA_CAUUUGCCACUUUUU_CCAUGGAUCGUACUUUUCG_GGCAUGGCAAAUGGCAGCGCUGAUUUACUU ....................................((((((.(((...(((((.................))))).)))))))))............. (-11.53 = -11.31 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:47 2006