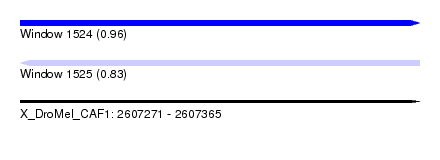
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,607,271 – 2,607,365 |
| Length | 94 |
| Max. P | 0.961444 |

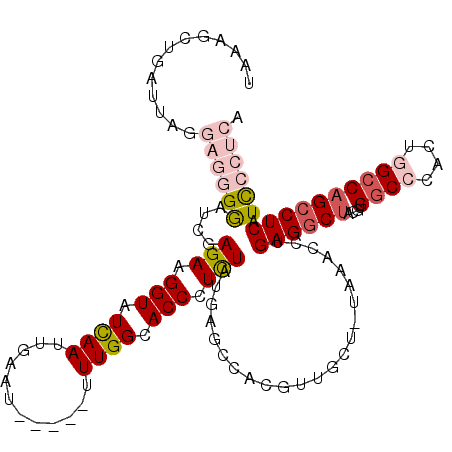
| Location | 2,607,271 – 2,607,365 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -17.74 |
| Energy contribution | -19.46 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2607271 94 + 22224390 UAAAGCUGAUUAGGAGGGGGUCGAGAAGGUAUCAAUUGAGU-----UUUGGCACCCUCUAU-------------------CUGAGGCUCCGUGGCCCACUGGCCAGCCUCAUUCCUCA .......(((..((((((.(((((((.....((....)).)-----)))))).))))))))-------------------)(((((((....((((....)))))))))))....... ( -34.40) >DroSec_CAF1 15235 110 + 1 UAAAGCUGAUUAG---GGAAUCGAGAAGGUAUCAAUUGAAU-----UUUGGCACCCUCUAUGAGCCACGUUGCUUUAAACCGGAGGCUACGUGGCCCACUGGCCAGCCUCAUCCCUCA ....((((..(((---((..((((((.(((..(((......-----.)))..))).))).)))(((((((.(((((......))))).))))))))).)))..))))........... ( -38.60) >DroSim_CAF1 14913 110 + 1 UAAAGCUGAUUAG---GGAAUCGAGAAGGUAUCAAUUGAAU-----UUUGGCACCCUCUAUGAGCCACGUUGCUUUAAACCGGAGGCUACGUGGCCCACUGGCCAGCCUCAUCCCUCA ....((((..(((---((..((((((.(((..(((......-----.)))..))).))).)))(((((((.(((((......))))).))))))))).)))..))))........... ( -38.60) >DroEre_CAF1 16227 115 + 1 UAAAGCUGAUUAGGAG-GGUGAGAGAAGGUAUCAAUUGAUACGAGUAUUGGUACCGUUUAUGAGGUACGUUGCC-UAAAUCUGAGGCUACGUGGC-UACUGGCCAGACUCAUUCCUCA .............(((-((((((....(((((((((..........)))))))))(((....((.(((((.(((-(.......)))).))))).)-)...)))....)))).))))). ( -37.30) >DroYak_CAF1 16580 110 + 1 UAAAGCUGAUUAGGAG-GGGACUAGAAGGUAUUAAUUGAUA-----AUUGGUACCCUUUAUGGGCUACGUUGCU-UAAAUCUGAGGCUCCGUGGC-UACUGGGCCGCCUCAUCCCUCA .............(((-(((..((((.(((((((((.....-----))))))))).))))(((((......)))-))....((((((.....(((-(....)))))))))))))))). ( -36.10) >consensus UAAAGCUGAUUAGGAGGGGAUCGAGAAGGUAUCAAUUGAAU_____UUUGGCACCCUCUAUGAGCCACGUUGCU_UAAACCUGAGGCUACGUGGCCCACUGGCCAGCCUCAUCCCUCA .............((((((....(((.(((.((((............)))).))).))).......................((((((....((((....)))))))))).)))))). (-17.74 = -19.46 + 1.72)

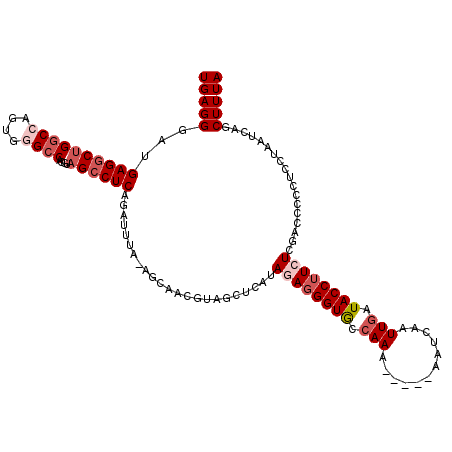
| Location | 2,607,271 – 2,607,365 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -17.48 |
| Energy contribution | -19.04 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2607271 94 - 22224390 UGAGGAAUGAGGCUGGCCAGUGGGCCACGGAGCCUCAG-------------------AUAGAGGGUGCCAAA-----ACUCAAUUGAUACCUUCUCGACCCCCUCCUAAUCAGCUUUA .((((..(((((((((((....))))....))))))).-------------------..((((((((.(((.-----......))).))))))))......))))............. ( -32.90) >DroSec_CAF1 15235 110 - 1 UGAGGGAUGAGGCUGGCCAGUGGGCCACGUAGCCUCCGGUUUAAAGCAACGUGGCUCAUAGAGGGUGCCAAA-----AUUCAAUUGAUACCUUCUCGAUUCC---CUAAUCAGCUUUA .......(((((((((...(((((((((((.((............)).)))))))))))((((((((.(((.-----......))).)))))))).......---....))))))))) ( -39.20) >DroSim_CAF1 14913 110 - 1 UGAGGGAUGAGGCUGGCCAGUGGGCCACGUAGCCUCCGGUUUAAAGCAACGUGGCUCAUAGAGGGUGCCAAA-----AUUCAAUUGAUACCUUCUCGAUUCC---CUAAUCAGCUUUA .......(((((((((...(((((((((((.((............)).)))))))))))((((((((.(((.-----......))).)))))))).......---....))))))))) ( -39.20) >DroEre_CAF1 16227 115 - 1 UGAGGAAUGAGUCUGGCCAGUA-GCCACGUAGCCUCAGAUUUA-GGCAACGUACCUCAUAAACGGUACCAAUACUCGUAUCAAUUGAUACCUUCUCUCACC-CUCCUAAUCAGCUUUA .((((...((((.((((.....-)))).((.((((.......)-))).))(((((........)))))....))))(((((....)))))..........)-)))............. ( -29.10) >DroYak_CAF1 16580 110 - 1 UGAGGGAUGAGGCGGCCCAGUA-GCCACGGAGCCUCAGAUUUA-AGCAACGUAGCCCAUAAAGGGUACCAAU-----UAUCAAUUAAUACCUUCUAGUCCC-CUCCUAAUCAGCUUUA .(((((.(((((((((......-))).....))))))((((.(-((.......((((.....))))....((-----((.....))))..)))..))))))-)))............. ( -27.40) >consensus UGAGGGAUGAGGCUGGCCAGUGGGCCACGUAGCCUCAGAUUUA_AGCAACGUAGCUCAUAGAGGGUGCCAAA_____AAUCAAUUGAUACCUUCUCGACCCCCUCCUAAUCAGCUUUA (((((...((((((((((....))))....)))))).......................((((((((.(((............))).))))))))..................))))) (-17.48 = -19.04 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:37 2006