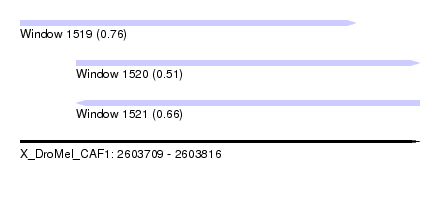
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,603,709 – 2,603,816 |
| Length | 107 |
| Max. P | 0.764893 |

| Location | 2,603,709 – 2,603,799 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -15.89 |
| Energy contribution | -17.39 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2603709 90 + 22224390 UCUU---CUUGUCGCAGCGGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCUCAGCGCCUUGGCCCAGC-CCAA ....---...(.(((((.((((((..((((........((((((.......))))))))))..)))))).))..))).)..(((...))-)... ( -25.20) >DroVir_CAF1 15303 82 + 1 CCUUGUCGCUGGCGCAGCGGAUGCUUGGCAUUUAUUGCAAGCGACAAUAUUUCGCUG---GCUGCAUCAGC--A---A--UG-GAGCAGCAGG- ..((((((((...((((..(((((...)))))..)))).)))))))).......(((---.((((..((..--.---.--))-..))))))).- ( -34.50) >DroSec_CAF1 11807 89 + 1 UCUU---CUUGUCGCAGCGGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCCCAGCGCCUUGGCCCAGC-CCA- ....---...(.(((.(.((((((..((((........((((((.......))))))))))..)))))).)...))).)..(((...))-)..- ( -23.70) >DroEre_CAF1 12742 89 + 1 UCUU---CUUGUCGCAGCGGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCUCAGCGCCUUGGCAACGC-CCA- ....---.(((((((((..(((((...)))))..))))(((((.........(((........))).........))))).)))))...-...- ( -25.77) >DroYak_CAF1 12106 89 + 1 UCUU---CUUGUCGCAGCGGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCUCAGCGCCUUAGCAACGC-CCA- ....---......((((.((((((..((((........((((((.......))))))))))..)))))).))..((......))...))-...- ( -24.00) >DroMoj_CAF1 19762 85 + 1 CCUUGUCGCUGGCGCAGCGGAUGCUUGGCAUUUAUUGCAAGCGACAAUAUUUCGCUG---GCUGCAUCAGC--AUCGC--UG-GAGCAGCAGG- ..((((((((...((((..(((((...)))))..)))).)))))))).......(((---.((((..((((--...))--))-..))))))).- ( -37.30) >consensus UCUU___CUUGUCGCAGCGGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCUCAGCGCCUUGGCAACGC_CCA_ .............((((..(((((...)))))..))))(((((.........(((........))).........))))).............. (-15.89 = -17.39 + 1.50)

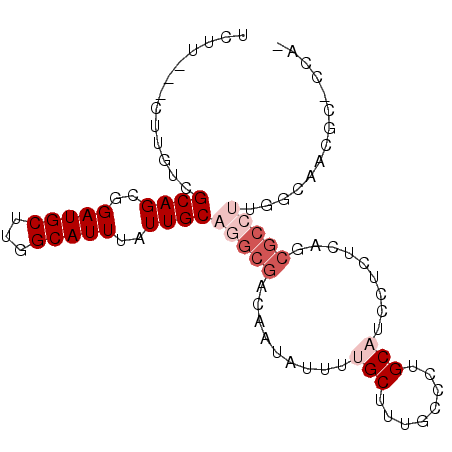
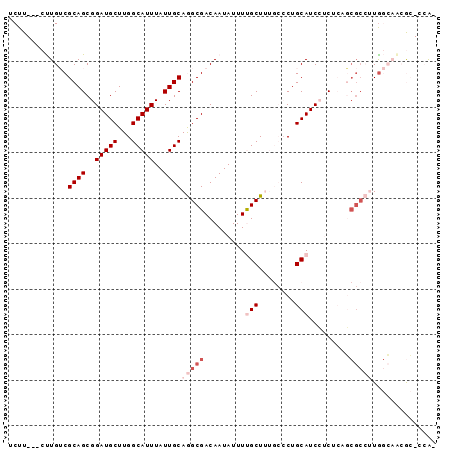
| Location | 2,603,724 – 2,603,816 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 83.78 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2603724 92 + 22224390 GGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCUCAGCGCCUUGGCCCAGC-CCAAACCACCUACCCCGCCCC ((((((..((((........((((((.......))))))))))..)))))).....(((....((......-........)).....)))... ( -21.34) >DroSec_CAF1 11822 87 + 1 GGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCCCAGCGCCUUGGCCCAGC-CCA-----CCGCCCCGCCCCC ((((((..((((........((((((.......))))))))))..)))))).....(((....(((...))-)..-----.)))......... ( -24.30) >DroEre_CAF1 12757 87 + 1 GGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCUCAGCGCCUUGGCAACGC-CCA-----CCGCCCCGCCCCU ((((((..((((........((((((.......))))))))))..)))))).....(((....(((.....-...-----..))).))).... ( -24.40) >DroYak_CAF1 12121 87 + 1 GGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCUCAGCGCCUUAGCAACGC-CCA-----CCGCCCCGCCUUU ((((((..((((........((((((.......))))))))))..)))))).....((((....))...((-...-----..))...)).... ( -21.60) >DroAna_CAF1 15518 77 + 1 GGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCU----------UGGGAGCACUCCC-----CCGCACCCUC-CC ((((((..((((........((((((.......))))))))))..)))))).----------.(((.((......-----..)).)))..-.. ( -25.90) >consensus GGAUGCUUGGCAUUUAUUGCAGGCGACAAUAUUUUGCUUUGCCCUGCAUCCUCUCAGCGCCUUGGCAACGC_CCA_____CCGCCCCGCCCCC ((((((..((((........((((((.......))))))))))..)))))).......................................... (-19.20 = -19.20 + -0.00)

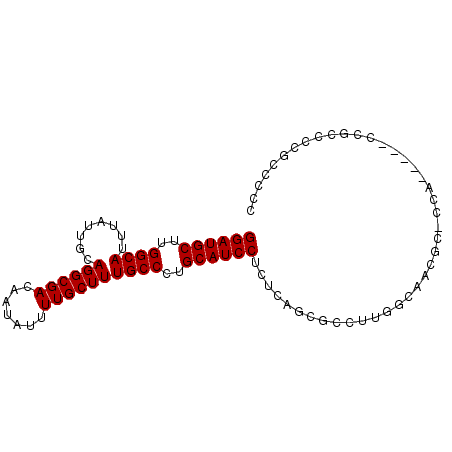
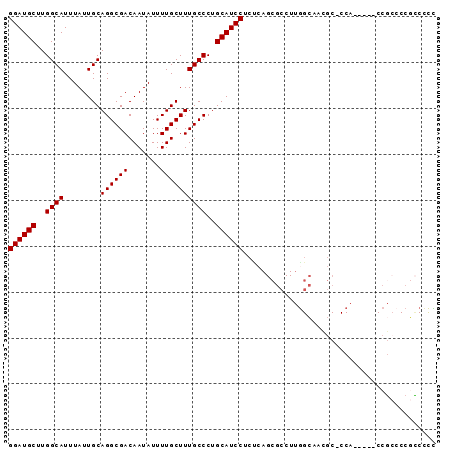
| Location | 2,603,724 – 2,603,816 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 83.78 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -19.96 |
| Energy contribution | -21.40 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2603724 92 - 22224390 GGGGCGGGGUAGGUGGUUUGG-GCUGGGCCAAGGCGCUGAGAGGAUGCAGGGCAAAGCAAAAUAUUGUCGCCUGCAAUAAAUGCCAAGCAUCC ..((((..((((((((.((..-(((..(((...(((((....)).)))..)))..)))..)).....))))))))......))))........ ( -32.90) >DroSec_CAF1 11822 87 - 1 GGGGGCGGGGCGG-----UGG-GCUGGGCCAAGGCGCUGGGAGGAUGCAGGGCAAAGCAAAAUAUUGUCGCCUGCAAUAAAUGCCAAGCAUCC ((..((..((((.-----...-(((..(((...(((((....)).)))..)))..)))....((((((.....))))))..))))..))..)) ( -29.60) >DroEre_CAF1 12757 87 - 1 AGGGGCGGGGCGG-----UGG-GCGUUGCCAAGGCGCUGAGAGGAUGCAGGGCAAAGCAAAAUAUUGUCGCCUGCAAUAAAUGCCAAGCAUCC .(((((..(((((-----..(-((((((((...(((((....)).)))..))))).((((....))))))))..)......))))..)).))) ( -33.70) >DroYak_CAF1 12121 87 - 1 AAAGGCGGGGCGG-----UGG-GCGUUGCUAAGGCGCUGAGAGGAUGCAGGGCAAAGCAAAAUAUUGUCGCCUGCAAUAAAUGCCAAGCAUCC ....((..(((((-----..(-((((((((...(((((....)).)))..))))).((((....))))))))..)......))))..)).... ( -30.20) >DroAna_CAF1 15518 77 - 1 GG-GAGGGUGCGG-----GGGAGUGCUCCCA----------AGGAUGCAGGGCAAAGCAAAAUAUUGUCGCCUGCAAUAAAUGCCAAGCAUCC ..-..((((((((-----(((....))))).----------....((((((.(...((((....)))).)))))))...........)))))) ( -27.10) >consensus GGGGGCGGGGCGG_____UGG_GCGGUGCCAAGGCGCUGAGAGGAUGCAGGGCAAAGCAAAAUAUUGUCGCCUGCAAUAAAUGCCAAGCAUCC ...((((((((...........))))))))............((((((.((((...((((....)))).))))(((.....)))...)))))) (-19.96 = -21.40 + 1.44)

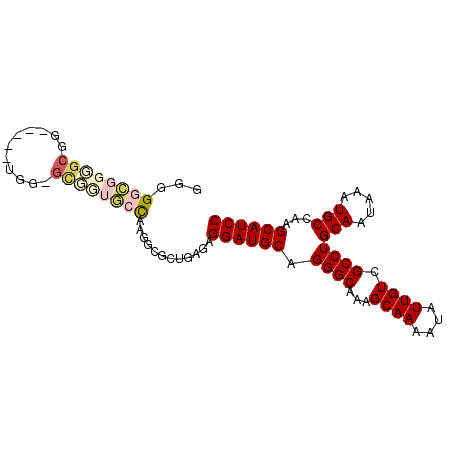
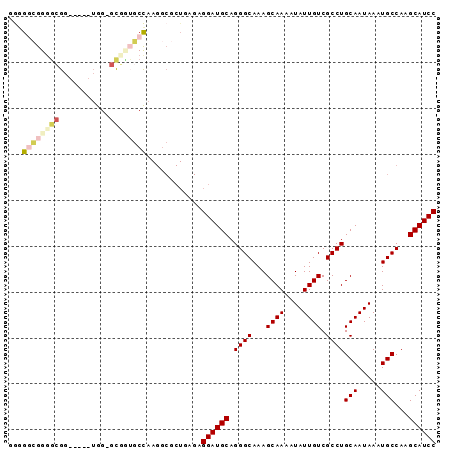
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:34 2006