| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,594,974 – 2,595,083 |
| Length | 109 |
| Max. P | 0.569418 |

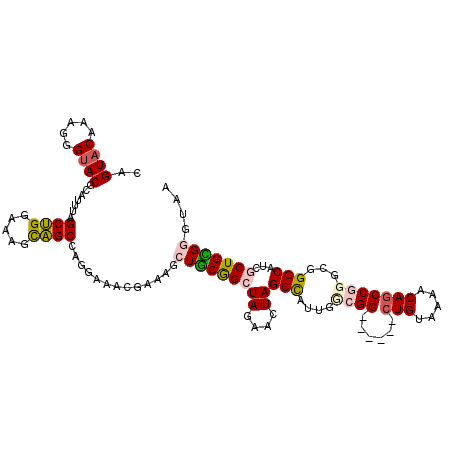
| Location | 2,594,974 – 2,595,083 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2594974 109 + 22224390 UUACCCGCAGCGAUGCCCGCUCGGUUGUUUUACAG------CCGCCAAUGGCUAGUUCUAGCCGCAGCUUUCGUUUCCUGGCUGCUUUCCAGCUAAAUGCGUACCCUUUGAACUA .....((((((((.((..((.((((((.....)))------))).....(((((....))))))).))..))))....((((((.....))))))..)))).............. ( -33.30) >DroSec_CAF1 3791 109 + 1 UUACUCGCAGCGAUGCCCGCCCGGCUGUUUUACAG------CCGACAAUGGCUAGUUCUAGCCGCAGAUUUCGUUUCCUGGCUGCUUUCCAGCUAAAUGCGUACCCUUUGUACUG ......(((((((...(.((.((((((.....)))------))).....(((((....))))))).)...))))....((((((.....))))))..)))((((.....)))).. ( -35.20) >DroSim_CAF1 3781 109 + 1 UUACUCGCAGCGAUGCCCGCCCGGCUGUUUUACAG------CCGACAAUGGCUAGUUCUAGCCGCAGCUUUCGUUUCCUGGCUGCUUUCCAGCUAAAUGCGUACCCUCUGUACUG ......(((((((.((..((.((((((.....)))------))).....(((((....))))))).))..))))....((((((.....))))))..)))((((.....)))).. ( -37.30) >DroEre_CAF1 4424 109 + 1 UUACCCACAGUGAUGCCCGCCAGGCUGUUUUCCAG------CCGGCAAUGGCUAGUUCUACCUGCAGCUUUCGUUUCUUGGCUGUUUUCCAGCUAAAUGAGUACGUUUUGUACUG .......((((...(((.(((.(((((.....)))------)))))...)))...........((((....(((((((((((((.....)))))))..))).)))..)))))))) ( -30.90) >DroYak_CAF1 3874 115 + 1 UUACCCUCAGUAAUGCCCGCUGGGCUGCUUACCAGUAAGUACCACUAAUAGCUAGUUCUAGCUGAAGCUUUCGUUACUUGGCGAUUUUCCCGCUAAAUGCGUACUUAUUGUACUG .......(((((..((((...)))).......((((((((((......((((((....))))))........((...((((((.......))))))..))))))))))))))))) ( -33.70) >consensus UUACCCGCAGCGAUGCCCGCCCGGCUGUUUUACAG______CCGACAAUGGCUAGUUCUAGCCGCAGCUUUCGUUUCCUGGCUGCUUUCCAGCUAAAUGCGUACCCUUUGUACUG ......((((((.....)))..(((((......................(((((....)))))((((((..........))))))....)))))...)))((((.....)))).. (-19.76 = -20.52 + 0.76)

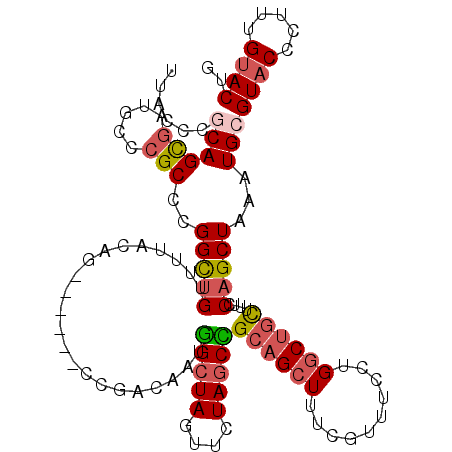
| Location | 2,594,974 – 2,595,083 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2594974 109 - 22224390 UAGUUCAAAGGGUACGCAUUUAGCUGGAAAGCAGCCAGGAAACGAAAGCUGCGGCUAGAACUAGCCAUUGGCGG------CUGUAAAACAACCGAGCGGGCAUCGCUGCGGGUAA ............(((.....((((((....(((((..(....)....)))))(((((....))))).....)))------)))........(((((((.....)))).)))))). ( -36.40) >DroSec_CAF1 3791 109 - 1 CAGUACAAAGGGUACGCAUUUAGCUGGAAAGCAGCCAGGAAACGAAAUCUGCGGCUAGAACUAGCCAUUGUCGG------CUGUAAAACAGCCGGGCGGGCAUCGCUGCGAGUAA ..((((.....))))((.....))......(((((..(....)((..((((((((((....)))))....((((------(((.....))))))))))))..)))))))...... ( -42.00) >DroSim_CAF1 3781 109 - 1 CAGUACAGAGGGUACGCAUUUAGCUGGAAAGCAGCCAGGAAACGAAAGCUGCGGCUAGAACUAGCCAUUGUCGG------CUGUAAAACAGCCGGGCGGGCAUCGCUGCGAGUAA ..((((.....))))(((....(((....))).(((.(....)....(((..(((((....))))).....(((------(((.....))))))))).))).....)))...... ( -40.70) >DroEre_CAF1 4424 109 - 1 CAGUACAAAACGUACUCAUUUAGCUGGAAAACAGCCAAGAAACGAAAGCUGCAGGUAGAACUAGCCAUUGCCGG------CUGGAAAACAGCCUGGCGGGCAUCACUGUGGGUAA .(((((.....)))))......((((.....)))).......(....)(..(((...((....(((...(((((------(((.....))))).))).))).)).)))..).... ( -34.50) >DroYak_CAF1 3874 115 - 1 CAGUACAAUAAGUACGCAUUUAGCGGGAAAAUCGCCAAGUAACGAAAGCUUCAGCUAGAACUAGCUAUUAGUGGUACUUACUGGUAAGCAGCCCAGCGGGCAUUACUGAGGGUAA (((((((.(((((((..((((.((((.....)))).)))).......(((..(((((....)))))...))).))))))).)).......((((...))))..)))))....... ( -31.40) >consensus CAGUACAAAGGGUACGCAUUUAGCUGGAAAGCAGCCAGGAAACGAAAGCUGCGGCUAGAACUAGCCAUUGGCGG______CUGUAAAACAGCCGGGCGGGCAUCGCUGCGGGUAA ..((((.....)))).......((((.....)))).............(((((((((....))(((....((((......(((.....)))))))...)))...))))))).... (-18.72 = -19.32 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:28 2006