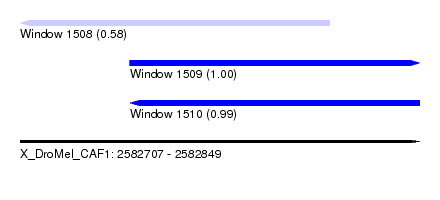
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,582,707 – 2,582,849 |
| Length | 142 |
| Max. P | 0.999915 |

| Location | 2,582,707 – 2,582,817 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -14.16 |
| Energy contribution | -15.64 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2582707 110 - 22224390 ACAGAAUCAAUAGAUAGAGUUGGCAGCUCUGAAUGAGACGAGCUAAGCUCC-UGGACCUGCCAACUCCUG--------CUGCGCAUAACGGUUAAUGCAAGACCAAACAUUUAUUAAAC ........(((((((.(((((((((((((.....)))..((((...)))).-.....))))))))))...--------....((((........))))..........))))))).... ( -29.40) >DroSec_CAF1 9796 110 - 1 ACAGAAUCAAUAGAUCGAGUUGGCAGCUCUAAAUGAGACAAGCACAGCUCC-UCAACCUGCCAACUCCUG--------CUGCGCAUAACGGUUAAUGCAAGACCAAACAUUUAUUAAAC ........(((((((.(((((((((((((.....)))...(((...)))..-.....))))))))))...--------....((((........))))..........))))))).... ( -25.30) >DroSim_CAF1 13159 110 - 1 ACAGAAUCAAUAGAUCGAGUUGGCAGCUCUAAUUGAGACGAGCACAGCUCC-UGAACCUGCCAACUCCUG--------CUGCGCAUAACGGUUAAUGCAAGACCAAACAUUUAUUAAAC ........(((((((.(((((((((((((.....)))..((((...)))).-.....))))))))))...--------....((((........))))..........))))))).... ( -28.90) >DroEre_CAF1 10123 108 - 1 ACAGGAUCGAUGCAACGAGUUGGCAGCUCUGAAGCAGACGUC---CGCUCUGCGAACCUGCCAACUGCUG--------CUGCGCAUAACGGUUCAUGCUGGACAAAACAUUUAUUUAUU ...((((((((((....(((((((((.......(((((.(..---..))))))....)))))))))((..--------..))))))..))))))......................... ( -32.90) >DroYak_CAF1 9990 107 - 1 ACAGGAUCAAUGAAACGAGUGGACAGCUCUGGACCUAACC------------AGGACCUGCCAACUCCUGCACUUCUGCACCACAUAACGGUUCUUGCUUGACAAAACAUUUAUUUAUU ..((((((........((((((.(((.(((((......))------------)))..))))).)))).((((....)))).........))))))........................ ( -22.30) >consensus ACAGAAUCAAUAGAUCGAGUUGGCAGCUCUGAAUGAGACGAGC__AGCUCC_UGAACCUGCCAACUCCUG________CUGCGCAUAACGGUUAAUGCAAGACCAAACAUUUAUUAAAC ........((((((..((((((((((........(((..........))).......))))))))))...............((((........))))...........)))))).... (-14.16 = -15.64 + 1.48)

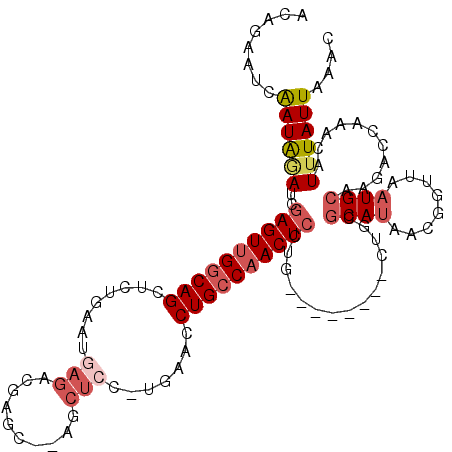
| Location | 2,582,746 – 2,582,849 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.95 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -22.81 |
| Energy contribution | -23.75 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2582746 103 + 22224390 AGCAGGAGUUGGCAGGUCCA-GGAGCUUAGCUCGUCUCAUUCAGAGCUGCCAACUCUAUCUAUUGAUUCUGUUAUUAAUUAGUUGCGCA--------CAAGAUCGAAAAGCU (((.(((((((((((.....-.((((...))))..(((.....)))))))))))))).....(((((..(((...(((....)))...)--------))..)))))...))) ( -32.00) >DroSec_CAF1 9835 103 + 1 AGCAGGAGUUGGCAGGUUGA-GGAGCUGUGCUUGUCUCAUUUAGAGCUGCCAACUCGAUCUAUUGAUUCUGUUGUUAAUUAGAAGCGCA--------AAAAAUCGAAAAGCU (((..((((((((((.((((-.((((...)))).)).......)).))))))))))(((.(.(((.(((((........)))))...))--------).).))).....))) ( -31.51) >DroSim_CAF1 13198 103 + 1 AGCAGGAGUUGGCAGGUUCA-GGAGCUGUGCUCGUCUCAAUUAGAGCUGCCAACUCGAUCUAUUGAUUCUGUUAUUAAUUAGAAGCGCA--------AAAGAUCGAAAAGCU (((..((((((((((.....-.((((...))))..(((.....)))))))))))))(((((.(((.(((((........)))))...))--------).))))).....))) ( -38.30) >DroEre_CAF1 10162 100 + 1 AGCAGCAGUUGGCAGGUUCGCAGAGCG---GACGUCUGCUUCAGAGCUGCCAACUCGUUGCAUCGAUCCUGUUAAUAACUUCAAUCGAAUUUAUGUUGUGCAU--------- .((((((((((((((.(((...(((((---(....))))))..)))))))))))).))))).(((((...((.....))....)))))...............--------- ( -36.70) >consensus AGCAGGAGUUGGCAGGUUCA_GGAGCUGUGCUCGUCUCAUUCAGAGCUGCCAACUCGAUCUAUUGAUUCUGUUAUUAAUUAGAAGCGCA________AAAGAUCGAAAAGCU (((((((((((((((.((....((((.......).))).....)).))))))))))((((....)))))))))....................................... (-22.81 = -23.75 + 0.94)

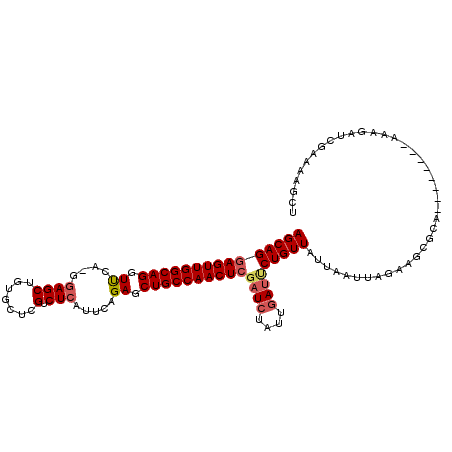
| Location | 2,582,746 – 2,582,849 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.95 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -16.78 |
| Energy contribution | -18.16 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2582746 103 - 22224390 AGCUUUUCGAUCUUG--------UGCGCAACUAAUUAAUAACAGAAUCAAUAGAUAGAGUUGGCAGCUCUGAAUGAGACGAGCUAAGCUCC-UGGACCUGCCAACUCCUGCU (((.....(((..((--------(................)))..)))........(((((((((((((.....)))..((((...)))).-.....))))))))))..))) ( -28.09) >DroSec_CAF1 9835 103 - 1 AGCUUUUCGAUUUUU--------UGCGCUUCUAAUUAACAACAGAAUCAAUAGAUCGAGUUGGCAGCUCUAAAUGAGACAAGCACAGCUCC-UCAACCUGCCAACUCCUGCU (((.....(((((.(--------((...((((..........)))).))).)))))(((((((((((((.....)))...(((...)))..-.....))))))))))..))) ( -26.00) >DroSim_CAF1 13198 103 - 1 AGCUUUUCGAUCUUU--------UGCGCUUCUAAUUAAUAACAGAAUCAAUAGAUCGAGUUGGCAGCUCUAAUUGAGACGAGCACAGCUCC-UGAACCUGCCAACUCCUGCU (((.....(((((.(--------((...((((..........)))).))).)))))(((((((((((((.....)))..((((...)))).-.....))))))))))..))) ( -32.20) >DroEre_CAF1 10162 100 - 1 ---------AUGCACAACAUAAAUUCGAUUGAAGUUAUUAACAGGAUCGAUGCAACGAGUUGGCAGCUCUGAAGCAGACGUC---CGCUCUGCGAACCUGCCAACUGCUGCU ---------...............((((((...((.....))..)))))).(((.(.(((((((((.......(((((.(..---..))))))....)))))))))).))). ( -27.80) >consensus AGCUUUUCGAUCUUU________UGCGCUUCUAAUUAAUAACAGAAUCAAUAGAUCGAGUUGGCAGCUCUAAAUGAGACGAGCACAGCUCC_UGAACCUGCCAACUCCUGCU ........(((((......................................)))))(((((((((((((.....)))..((((...)))).......))))))))))..... (-16.78 = -18.16 + 1.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:23 2006