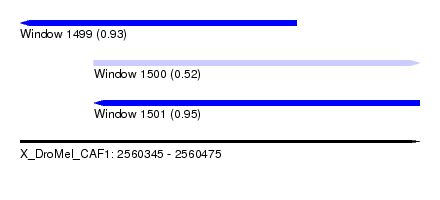
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,560,345 – 2,560,475 |
| Length | 130 |
| Max. P | 0.952981 |

| Location | 2,560,345 – 2,560,435 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 68.10 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -9.65 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2560345 90 - 22224390 UUCGCUGGGCUUAAAAAUUUAACAUACAUGUGCAUGUAGAACAAGCAUGCACAAUAUGUAUUUAUG-----------------UAUUUCGUCCAUGAGUUU----UCUUGU ((((.(((((..(((.((.(((.((((((((((((((.......)))))))))...)))))))).)-----------------).))).)))))))))...----...... ( -22.30) >DroSec_CAF1 2366 104 - 1 UCCGCUGAGCUAAAACAUUUAACAUACAUGUGUAUGUAAAACAAUCAUACACAAUAUGUAUUUACGCUCAUAGA------UGUUAGUUCGUCUAUUAGUUU-UUCUCUAGU ...((((((..(((((.....(((((..((((((((.........)))))))).)))))..........(((((------((......)))))))..))))-).))).))) ( -24.80) >DroYak_CAF1 3964 110 - 1 UCCGCUGAAUUAAAAAAUGUAACAUACAAAUGCAUGUAGAACAAACAUGUGCA-CAUGCAUGCAUAUUCUUACAUCUACCUGUUAGUUCGCCAAUAAAUUUUUUCUCUAGU ...((.(((((((...((((((.(((...((((((((...(((....)))..)-)))))))...)))..)))))).......))))))))).................... ( -20.60) >consensus UCCGCUGAGCUAAAAAAUUUAACAUACAUGUGCAUGUAGAACAAACAUGCACAAUAUGUAUUUAUG_UC_UA_A______UGUUAGUUCGUCAAUAAGUUU_UUCUCUAGU ...((.(((((((...............(((((((((.......)))))))))..(((....))).................))))))))).................... ( -9.65 = -10.00 + 0.35)

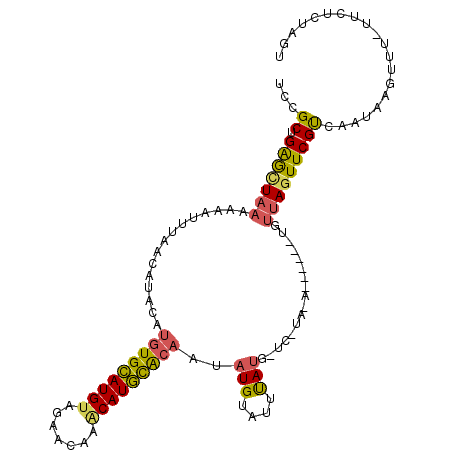
| Location | 2,560,369 – 2,560,475 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.84 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -5.58 |
| Energy contribution | -6.76 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.23 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2560369 106 + 22224390 --------CAUAAAUACAUAUUGUGCAUGCUUGUUCUACAUGCACAUGUAUGUUAAAUUUU------UAAGCCCAGCGAAAUUUAUUUUUGCCGCACUAGGUUGGCUAUUUACUCAGCUG --------.......((((((((((((((.........)))))))).))))))........------...((...((((((.....)))))).))....((((((........)))))). ( -24.40) >DroSec_CAF1 2396 114 + 1 UCUAUGAGCGUAAAUACAUAUUGUGUAUGAUUGUUUUACAUACACAUGUAUGUUAAAUGUU------UUAGCUCAGCGGAAUUUAUUUAUGCCGCACUAGGUUGGAUAUUUACUCAACUG ....(((((..((((((((((((((((((.........)))))))).)))))).....)))------)..)))))((((.((......)).))))....((((((........)))))). ( -30.10) >DroSim_CAF1 4451 86 + 1 -----AAGCGUAAAUAAAUAUUGUGC-----------------------AUGUUAAAUGUU------UUAGCUCAGCGGAAUUUAUUUAUGCCGCACUAGGUUGGAUAUUUACUCAGCUG -----..((((((((((((.((((..-----------------------..(((((.....------)))))...)))).)))))))))))).......((((((........)))))). ( -20.90) >DroEre_CAF1 2569 105 + 1 UGUAUGAACAUGCA-------------UGUUUGUUCU--AUGCAUUUGUAUGAUAAAUGUUUUUGCUUUAGCUCAGCGGAACUUAUUUAUAUCGCACUAAGUUGGCUAUUUAUUCAGCUG ((((((((((....-------------....))))).--)))))..(((...(((((((.(((((((.......)))))))..)))))))...)))...((((((........)))))). ( -21.90) >DroYak_CAF1 4001 112 + 1 UGUAAGAAUAUGCAUGCAUG-UGCACAUGUUUGUUCUACAUGCAUUUGUAUGUUACAUUUU------UUAAUUCAGCGGAACAUAUUUAUAGC-CAUUGAGUUUGCUAUUUAUUCAGCUG (((((..(((((.(((((((-((.(((....)))..))))))))).))))).)))))....------......((((.((..(((...(((((-.(......).))))).))))).)))) ( -25.70) >consensus U_UA_GAACAUAAAUACAUAUUGUGCAUGUUUGUUCUACAUGCACAUGUAUGUUAAAUGUU______UUAGCUCAGCGGAAUUUAUUUAUGCCGCACUAGGUUGGCUAUUUACUCAGCUG .....................................((((((....))))))......................(((..............)))....((((((........)))))). ( -5.58 = -6.76 + 1.18)

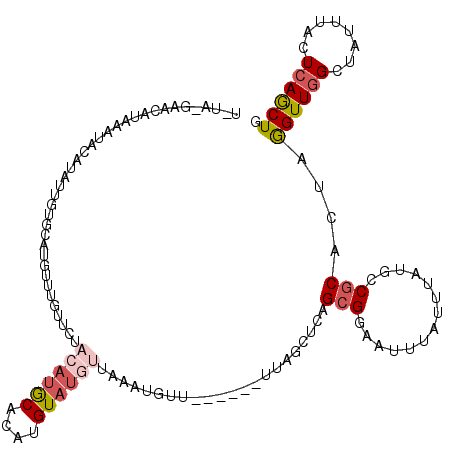
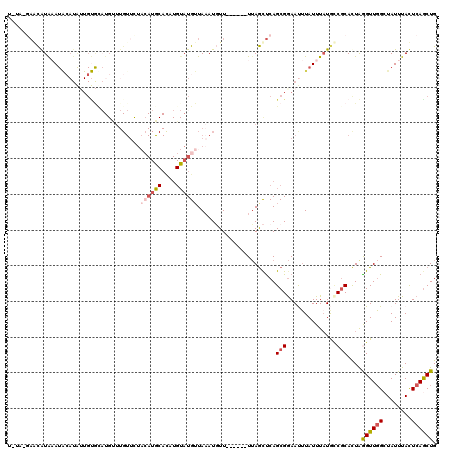
| Location | 2,560,369 – 2,560,475 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.84 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -5.83 |
| Energy contribution | -5.03 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.27 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
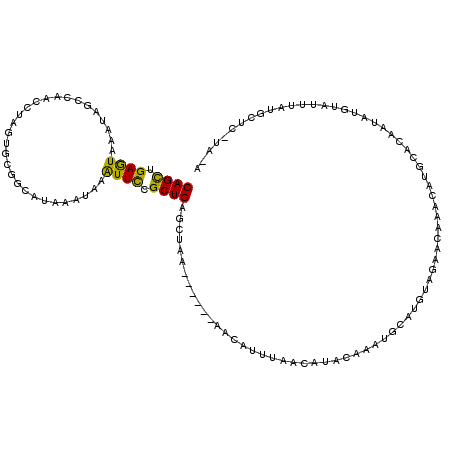
>X_DroMel_CAF1 2560369 106 - 22224390 CAGCUGAGUAAAUAGCCAACCUAGUGCGGCAAAAAUAAAUUUCGCUGGGCUUA------AAAAUUUAACAUACAUGUGCAUGUAGAACAAGCAUGCACAAUAUGUAUUUAUG-------- ..((((......))))......(((.((((.(((.....))).)))).)))..------........(((((..(((((((((.......))))))))).))))).......-------- ( -25.40) >DroSec_CAF1 2396 114 - 1 CAGUUGAGUAAAUAUCCAACCUAGUGCGGCAUAAAUAAAUUCCGCUGAGCUAA------AACAUUUAACAUACAUGUGUAUGUAAAACAAUCAUACACAAUAUGUAUUUACGCUCAUAGA ..((((..(....)..))))(((..((((.((......)).))))((((((((------(.......(((((..((((((((.........)))))))).))))).)))).)))))))). ( -24.10) >DroSim_CAF1 4451 86 - 1 CAGCUGAGUAAAUAUCCAACCUAGUGCGGCAUAAAUAAAUUCCGCUGAGCUAA------AACAUUUAACAU-----------------------GCACAAUAUUUAUUUACGCUU----- .((((((((((((((..........((((.((......)).))))((.((...------............-----------------------)).))))))))))))).))).----- ( -15.86) >DroEre_CAF1 2569 105 - 1 CAGCUGAAUAAAUAGCCAACUUAGUGCGAUAUAAAUAAGUUCCGCUGAGCUAAAGCAAAAACAUUUAUCAUACAAAUGCAU--AGAACAAACA-------------UGCAUGUUCAUACA ....((((((....((...(((((((.(((........))).))))))).....))....................(((((--.(......))-------------)))))))))).... ( -16.00) >DroYak_CAF1 4001 112 - 1 CAGCUGAAUAAAUAGCAAACUCAAUG-GCUAUAAAUAUGUUCCGCUGAAUUAA------AAAAUGUAACAUACAAAUGCAUGUAGAACAAACAUGUGCA-CAUGCAUGCAUAUUCUUACA ((((.(((((.(((((..........-))))).....))))).))))......------....(((((.(((...((((((((...(((....)))..)-)))))))...)))..))))) ( -24.90) >consensus CAGCUGAGUAAAUAGCCAACCUAGUGCGGCAUAAAUAAAUUCCGCUGAGCUAA______AACAUUUAACAUACAAAUGCAUGUAGAACAAACAUGCACAAUAUGUAUUUAUGCUC_UA_A ((((.((((.............................)))).))))......................................................................... ( -5.83 = -5.03 + -0.80)

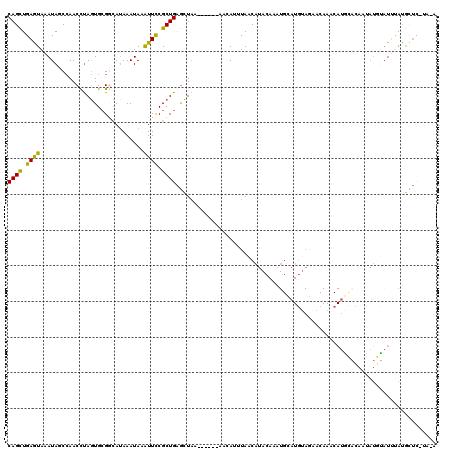
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:15 2006