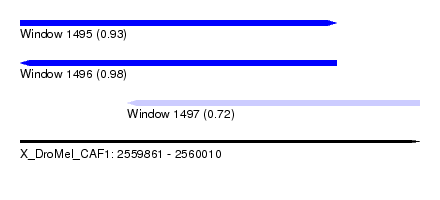
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,559,861 – 2,560,010 |
| Length | 149 |
| Max. P | 0.983457 |

| Location | 2,559,861 – 2,559,979 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -48.48 |
| Consensus MFE | -39.74 |
| Energy contribution | -41.10 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2559861 118 + 22224390 GACGACUCCACUGGACCGGGCGACACGUAAACAACAAUCAGAGCCCGAAGCUUCUGCCAGCGGCGGCU--UUUUAUCGCCGCCUAUGUGGUUGUUGUUGCUGCGCGCCAGGGACGUGGUC (((.(((((.((((.....(((.((.((((.(((((((((.(.......(((......)))((((((.--.......))))))..).)))))))))))))))))).))))))).)).))) ( -49.30) >DroSec_CAF1 1881 118 + 1 GACGACUCCACCGGACCGGGCGACACGUAAACAACAAUCAGAGCCCGAAGCUUCUGCCAGCGGCGGCU--UUUUAUCGCCGCCUAUGUGGUUGUUGUUGCUGUGCGCCAGGGACGUAGUC ...((((....((..((.((((.((((..(((((((((((.(.......(((......)))((((((.--.......))))))..).)))))))))))..)))))))).))..)).)))) ( -50.80) >DroSim_CAF1 3935 118 + 1 GACGACUCCACCGGACCGGGCGACACGUAAACAACAAUCAGAGCCCGAAGCUUCUGCCAGCGGCGGCU--UUUUAUCGCCGCCUAUGUGGUUGUUGUUGCUGUGCGCCAGGGACGUAGUC ...((((....((..((.((((.((((..(((((((((((.(.......(((......)))((((((.--.......))))))..).)))))))))))..)))))))).))..)).)))) ( -50.80) >DroEre_CAF1 2063 118 + 1 GACGACUCCACCGGACCGGGCGACACGUAAACAACAAUCAGAGCCCGAAGCUUCUGCCAGCGGCGA-UUUUUUUAUCGCCGCCAAUGUGGUUGUUGUUGCUGUG-GCCAUGGGCGUGGCC .......((((....((((((.(((.((((.(((((((((((((.....))))......(((((((-(......)))))))).....)))))))))))))))).-))).)))..)))).. ( -48.30) >DroYak_CAF1 3422 119 + 1 GACGACUCCACCGGACCGGUCGACACGUAAACAACAAUCAGAGCCCGAAGCUUCUGCCAGCGGCGACUUUUUUUAUCGCCGCCAAUGUGGUUGUUGUUGCUGUG-GCCAUGGGCGUGGCC ..(((((((...)))...))))(((.((((.(((((((((((((.....))))......(((((((.........))))))).....))))))))))))))))(-(((((....)))))) ( -43.20) >consensus GACGACUCCACCGGACCGGGCGACACGUAAACAACAAUCAGAGCCCGAAGCUUCUGCCAGCGGCGGCU__UUUUAUCGCCGCCUAUGUGGUUGUUGUUGCUGUGCGCCAGGGACGUGGUC .......((((.(..((.((((.((((..(((((((((((((((.....))))......(((((((.........))))))).....)))))))))))..)))))))).))..))))).. (-39.74 = -41.10 + 1.36)

| Location | 2,559,861 – 2,559,979 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -49.50 |
| Consensus MFE | -40.20 |
| Energy contribution | -40.76 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2559861 118 - 22224390 GACCACGUCCCUGGCGCGCAGCAACAACAACCACAUAGGCGGCGAUAAAA--AGCCGCCGCUGGCAGAAGCUUCGGGCUCUGAUUGUUGUUUACGUGUCGCCCGGUCCAGUGGAGUCGUC ..(((((..((.(((((((...(((((((((((....((((((.......--.))))))..)))((((.((.....)))))).))))))))...))).)))).))..).))))....... ( -50.50) >DroSec_CAF1 1881 118 - 1 GACUACGUCCCUGGCGCACAGCAACAACAACCACAUAGGCGGCGAUAAAA--AGCCGCCGCUGGCAGAAGCUUCGGGCUCUGAUUGUUGUUUACGUGUCGCCCGGUCCGGUGGAGUCGUC ((((.((..((.(((((((...(((((((((((....((((((.......--.))))))..)))((((.((.....)))))).))))))))...))).)))).))..))....))))... ( -52.10) >DroSim_CAF1 3935 118 - 1 GACUACGUCCCUGGCGCACAGCAACAACAACCACAUAGGCGGCGAUAAAA--AGCCGCCGCUGGCAGAAGCUUCGGGCUCUGAUUGUUGUUUACGUGUCGCCCGGUCCGGUGGAGUCGUC ((((.((..((.(((((((...(((((((((((....((((((.......--.))))))..)))((((.((.....)))))).))))))))...))).)))).))..))....))))... ( -52.10) >DroEre_CAF1 2063 118 - 1 GGCCACGCCCAUGGC-CACAGCAACAACAACCACAUUGGCGGCGAUAAAAAAA-UCGCCGCUGGCAGAAGCUUCGGGCUCUGAUUGUUGUUUACGUGUCGCCCGGUCCGGUGGAGUCGUC (((((......))))-).....((((((((.....(..((((((((......)-)))))))..)((((.((.....)))))).)))))))).(((..(((((......)))))...))). ( -45.90) >DroYak_CAF1 3422 119 - 1 GGCCACGCCCAUGGC-CACAGCAACAACAACCACAUUGGCGGCGAUAAAAAAAGUCGCCGCUGGCAGAAGCUUCGGGCUCUGAUUGUUGUUUACGUGUCGACCGGUCCGGUGGAGUCGUC (((((......))))-).....((((((((.....(..((((((((.......))))))))..)((((.((.....)))))).)))))))).(((..((.((((...)))).))..))). ( -46.90) >consensus GACCACGUCCCUGGCGCACAGCAACAACAACCACAUAGGCGGCGAUAAAA__AGCCGCCGCUGGCAGAAGCUUCGGGCUCUGAUUGUUGUUUACGUGUCGCCCGGUCCGGUGGAGUCGUC ..(((((..((.(((((((...(((((((((((....((((((..........))))))..)))((((.((.....)))))).))))))))...))).)))).))..).))))....... (-40.20 = -40.76 + 0.56)

| Location | 2,559,901 – 2,560,010 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.00 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
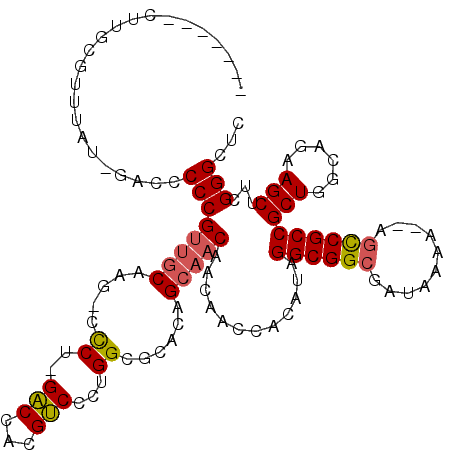
>X_DroMel_CAF1 2559901 109 - 22224390 -------UUCGCAUUUAU-GACCCCCGUUGCAAG-CCCAGGACCACGUCCCUGGCGCGCAGCAACAACAACCACAUAGGCGGCGAUAAAA--AGCCGCCGCUGGCAGAAGCUUCGGGCUC -------...((......-.......(((((..(-((((((........))))).))...))))).....((.....((((((.......--.))))))(((......)))...)))).. ( -40.80) >DroSec_CAF1 1921 108 - 1 -------CUUGCGUUUAU-AACCCCCGUUGCCAG-CCCU-GACUACGUCCCUGGCGCACAGCAACAACAACCACAUAGGCGGCGAUAAAA--AGCCGCCGCUGGCAGAAGCUUCGGGCUC -------...........-....(((((((((((-(...-......((..(((.....)))..))............((((((.......--.))))))))))))))......))))... ( -36.40) >DroSim_CAF1 3975 108 - 1 -------CUUGCGUUUAU-AACCCCCGUUGCCAG-CCCU-GACUACGUCCCUGGCGCACAGCAACAACAACCACAUAGGCGGCGAUAAAA--AGCCGCCGCUGGCAGAAGCUUCGGGCUC -------...........-....(((((((((((-(...-......((..(((.....)))..))............((((((.......--.))))))))))))))......))))... ( -36.40) >DroEre_CAF1 2103 116 - 1 AAGCAGGUUUGCCUUCAU-GGCCCCCGUGCCAAAACCCU-GGCCACGCCCAUGGC-CACAGCAACAACAACCACAUUGGCGGCGAUAAAAAAA-UCGCCGCUGGCAGAAGCUUCGGGCUC ((((...((((((....(-((((..(((((((......)-)).)))).....)))-))...................(((((((((......)-)))))))))))))).))))....... ( -42.50) >DroYak_CAF1 3462 112 - 1 ------GCUUGCGUUCAAGGGUCCCCGCUGCAAGACUCU-GGCCACGCCCAUGGC-CACAGCAACAACAACCACAUUGGCGGCGAUAAAAAAAGUCGCCGCUGGCAGAAGCUUCGGGCUC ------(((((.(((...((....)).((((..(.((.(-(((((......))))-)).)))...............(((((((((.......))))))))).)))).)))..))))).. ( -39.90) >consensus _______CUUGCGUUUAU_GACCCCCGUUGCAAG_CCCU_GACCACGUCCCUGGCGCACAGCAACAACAACCACAUAGGCGGCGAUAAAA__AGCCGCCGCUGGCAGAAGCUUCGGGCUC .......................((((((((.....((..(((...)))...))......)))))............((((((..........))))))(((......)))...)))... (-25.08 = -25.00 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:11 2006