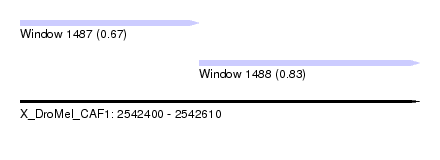
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,542,400 – 2,542,610 |
| Length | 210 |
| Max. P | 0.825631 |

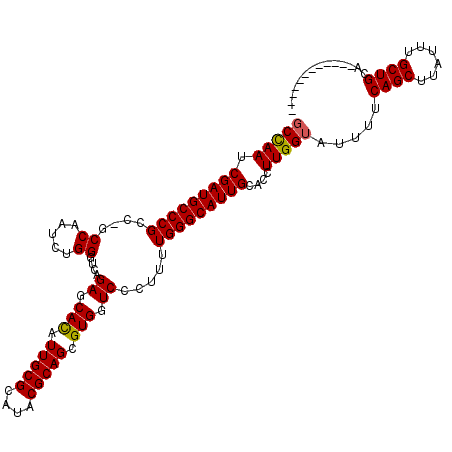
| Location | 2,542,400 – 2,542,494 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 59.01 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -11.94 |
| Energy contribution | -10.88 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2542400 94 + 22224390 ---CUGCAG---CCG-CUUGCCUUCCGGUCACCUGGAGUGCAGUACUG-CAGCACACUGCGCCUACGUGAAG-UGAACUCAUUCAUAUAUGUAGCUAC-AUAGC ---((((((---..(-((.((.((((((....)))))).))))).)))-)))..((((.(((....))).))-))............(((((....))-))).. ( -28.40) >DroSec_CAF1 5630 85 + 1 ---CUGCAGCCGCCG-CUUGCCUUCCGGUCACCUGGAGUGCAGCAC-----GCACACUGCGGCUACGUGGAG-UGAACUCAC--------AUAGCUGC-ACAAA ---.((((((.((((-(......(((((....)))))((((.....-----))))...)))))...((((((-....))).)--------)).)))))-).... ( -36.60) >DroSim_CAF1 5606 85 + 1 ---CUGCAGUCGCCG-CUUGCCUUCCGGUCACCUGGAGUGCAGCAC-----GCACACUGCGGCUACGUGGAG-UGAACUCAC--------AUAGCUUC-ACCUA ---..(((((.((.(-((.((.((((((....)))))).)))))..-----))..)))))(((((.(((.((-....)))))--------.)))))..-..... ( -31.50) >DroPer_CAF1 53167 88 + 1 UGUGUGUGG---CCGUUUUGCCCUUCGAACGUUUGAAA-GCUAUAAUGGCAG------------AGAUGAUGCCGUAUUCGUCUCUGUCUGUAGUCACCACGGC .(((.((((---(......((..(((((....))))).-))......(((((------------((((((........)))))))))))....))))))))... ( -30.00) >consensus ___CUGCAG___CCG_CUUGCCUUCCGGUCACCUGGAGUGCAGCAC_____GCACACUGCGGCUACGUGAAG_UGAACUCAC________AUAGCUAC_ACAGA .....(((((.......((((.((((((....)))))).)))).......................((((........))))...........)))))...... (-11.94 = -10.88 + -1.06)

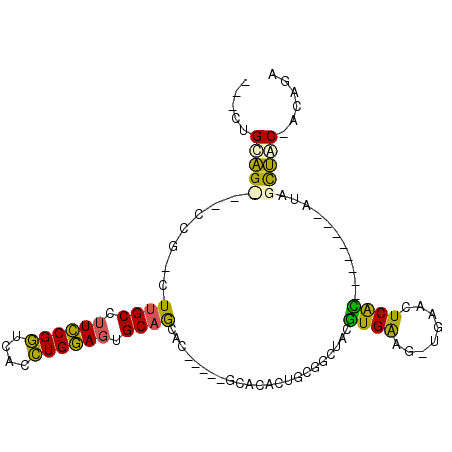
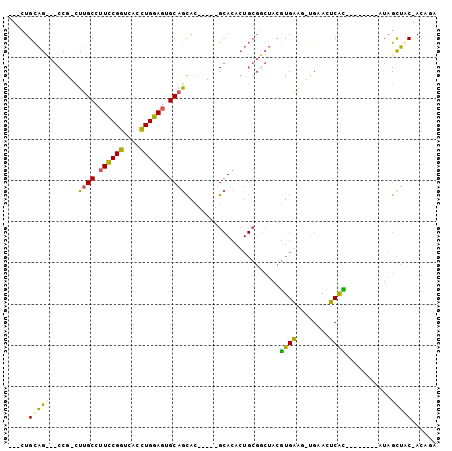
| Location | 2,542,494 – 2,542,610 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -32.37 |
| Energy contribution | -32.27 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2542494 116 + 22224390 UCUAAUCGAUGCCCGGCAUCCAAUCUGGGUCAGAGCAUAUUGCGCAUACGCAGCGUGUUCCCUUUUGGGCAUUGCACCUUGGUAUUUUCAGCUUAUUUGCUGCACACACGCCACGU ......((((((((((.((((.....))))).(((((..(((((....)))))..))))).....))))))))).....((((.....((((......)))).......))))... ( -36.80) >DroSec_CAF1 5715 103 + 1 GCCAAUCGAUGCCCGCC-GCCAAUCUGGGUCAGAGCACAUUGCGCAUACGCAGCGUGGUCCCUUUUGGGCAUUGCACCUUGGUAUUUUCAGCUUAUUUGCUGCA------------ (((((.((((((((...-(((......)))..((.(((.(((((....))))).))).))......))))))))....))))).....((((......))))..------------ ( -36.00) >DroSim_CAF1 5691 103 + 1 GCCAAUCGAUGCCCGCC-GCCAAUGUGGGUCAGAGCACAUUGCGCAUACGCAGCGUGGUCCCUUUUGGGCAUUGCACCUUGGUAUUUUCAGCUUAGUUGCUGCA------------ (((((.((((((((.((-((....))))....((.(((.(((((....))))).))).))......))))))))....))))).....((((......))))..------------ ( -38.20) >consensus GCCAAUCGAUGCCCGCC_GCCAAUCUGGGUCAGAGCACAUUGCGCAUACGCAGCGUGGUCCCUUUUGGGCAUUGCACCUUGGUAUUUUCAGCUUAUUUGCUGCA____________ (((((.(((((((((....((.....))....((.(((.(((((....))))).))).)).....)))))))))....))))).....((((......)))).............. (-32.37 = -32.27 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:02 2006