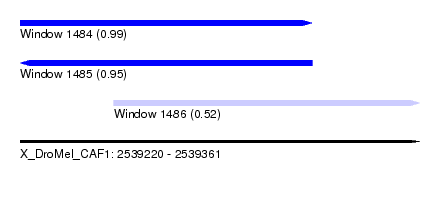
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,539,220 – 2,539,361 |
| Length | 141 |
| Max. P | 0.988398 |

| Location | 2,539,220 – 2,539,323 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.36 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -15.58 |
| Energy contribution | -16.97 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2539220 103 + 22224390 CCUUGUUUUC------CCCCUCGAAGCAGAUGUUAA-UCCCAACU-UGGGAUUAAGGUUC---UUCAAAAGUUUCGCGAAAGGCGUUGAAAUUUCAACGCCUUCGUCGUCCUAC ..........------......((((..(((.((((-((((....-.)))))))).))))---)))...((..(.(((((.((((((((....))))))))))))).)..)).. ( -35.60) >DroSec_CAF1 2587 103 + 1 CCUUGUUUCC------CCCCUCGCAGAAGAUGUUAA-UCCCAACU-UGGGAUUAUGGACC---CUCAAAAGUUUCGCGAAAGGCGUUGAAAUUUCAACGCCUCCGUCGUCUUAC ....(..(((------......(((.....)))(((-((((....-.))))))).)))..---)...........((((.(((((((((....)))))))))...))))..... ( -29.00) >DroSim_CAF1 2576 103 + 1 CCUUGUUUUC------CCCCUCGCAGAAGAUGUUAA-UCCCAACU-UGGGUUUAAGGACC---CUCAAAAGUUUCGCGAAAGGCGUUGAAAUUUCAACGCCUCCGUCGUCUUAC ..........------..........((((((....-....((((-((((((....))))---)....)))))..(((..(((((((((....))))))))).))))))))).. ( -27.10) >DroEre_CAF1 2759 101 + 1 CCUCGUUUUUUU---CACU-CC---GAAGAUGUUAA-ACCCAACU-UGGGAUUAAG-GUC---CGUCAAAGUUUCGCGAAAGGCGUUGAAAUUUCCACGCCUCCGUCGUCUUGC ............---.(((-..---((.(((.((((-.(((....-.))).)))).-)))---..))..)))...((((.((((((.((....)).))))))...))))..... ( -24.90) >DroYak_CAF1 2802 95 + 1 CCUUGUUUUUUA---CCCCCUC---GAAGAUGUUCA-ACUCAACU-UGGGAUUAAGGGUC---CUGCAAAGUUUCUCGAAAGGCGUUGAAAUUUCAACGCCUCCUC-------- ............---.....((---((.((......-....((((-((((((.....)))---))...))))))))))).(((((((((....)))))))))....-------- ( -21.90) >DroAna_CAF1 2409 102 + 1 CCUUAUUUUCUUUUCUUUU-CC---GGAAAUCUCGAGAACCAUCGCGGGGAUUAAUAUUUCCAGUUAAAAGUUUUCCUUAAGGCGUUGAAAUUUCAACGCUUUUUC-------- ...................-..---(((((.(((((......))).((..((....))..)).......)).)))))..((((((((((....))))))))))...-------- ( -23.00) >consensus CCUUGUUUUC______CCCCUC___GAAGAUGUUAA_ACCCAACU_UGGGAUUAAGGAUC___CUCAAAAGUUUCGCGAAAGGCGUUGAAAUUUCAACGCCUCCGUCGUCUUAC ......................................(((......))).........................((((.(((((((((....)))))))))...))))..... (-15.58 = -16.97 + 1.39)

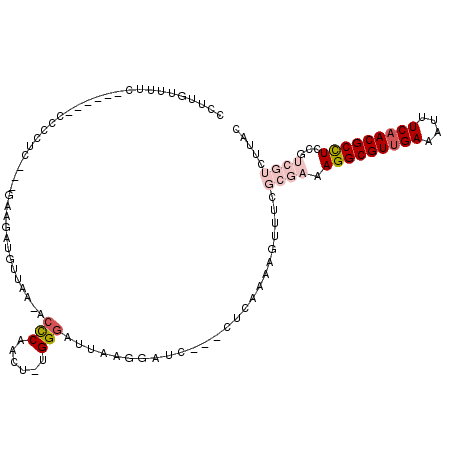
| Location | 2,539,220 – 2,539,323 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.36 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -15.04 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2539220 103 - 22224390 GUAGGACGACGAAGGCGUUGAAAUUUCAACGCCUUUCGCGAAACUUUUGAA---GAACCUUAAUCCCA-AGUUGGGA-UUAACAUCUGCUUCGAGGGG------GAAAACAAGG ......((.((((((((((((....)))))))).))))))...((((((((---(....((((((((.-....))))-))))......))))))))).------.......... ( -38.40) >DroSec_CAF1 2587 103 - 1 GUAAGACGACGGAGGCGUUGAAAUUUCAACGCCUUUCGCGAAACUUUUGAG---GGUCCAUAAUCCCA-AGUUGGGA-UUAACAUCUUCUGCGAGGGG------GGAAACAAGG ......((.((((((((((((....)))))))).))))))...((((.(((---(((...(((((((.-....))))-)))..))))))...))))..------(....).... ( -35.90) >DroSim_CAF1 2576 103 - 1 GUAAGACGACGGAGGCGUUGAAAUUUCAACGCCUUUCGCGAAACUUUUGAG---GGUCCUUAAACCCA-AGUUGGGA-UUAACAUCUUCUGCGAGGGG------GAAAACAAGG ......((.((((((((((((....)))))))).)))))).....((((((---....))))))(((.-....))).-......((((((....))))------))........ ( -31.00) >DroEre_CAF1 2759 101 - 1 GCAAGACGACGGAGGCGUGGAAAUUUCAACGCCUUUCGCGAAACUUUGACG---GAC-CUUAAUCCCA-AGUUGGGU-UUAACAUCUUC---GG-AGUG---AAAAAAACGAGG ......((.(((((((((.((....)).))))).))))))..(((((((.(---((.-.((((.(((.-....))).-))))..)))))---))-))).---............ ( -28.80) >DroYak_CAF1 2802 95 - 1 --------GAGGAGGCGUUGAAAUUUCAACGCCUUUCGAGAAACUUUGCAG---GACCCUUAAUCCCA-AGUUGAGU-UGAACAUCUUC---GAGGGGG---UAAAAAACAAGG --------(((.(((((((((....))))))))))))......(((((...---.((((((..((...-....)).(-((((....)))---)))))))---)......))))) ( -25.10) >DroAna_CAF1 2409 102 - 1 --------GAAAAAGCGUUGAAAUUUCAACGCCUUAAGGAAAACUUUUAACUGGAAAUAUUAAUCCCCGCGAUGGUUCUCGAGAUUUCC---GG-AAAAGAAAAGAAAAUAAGG --------......(((((((....)))))))...........(((((..((((((((........((.....))........))))))---))-))))).............. ( -20.49) >consensus GUAAGACGACGGAGGCGUUGAAAUUUCAACGCCUUUCGCGAAACUUUUGAG___GAACCUUAAUCCCA_AGUUGGGA_UUAACAUCUUC___GAGGGG______GAAAACAAGG ..........(((((((((((....)))))))))))............................(((......)))...................................... (-15.04 = -15.65 + 0.61)

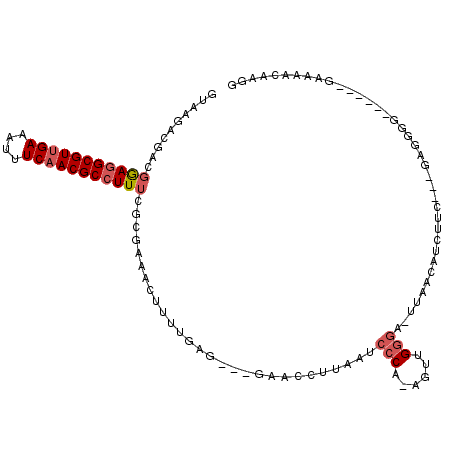
| Location | 2,539,253 – 2,539,361 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.35 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -11.11 |
| Energy contribution | -12.90 |
| Covariance contribution | 1.79 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.39 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
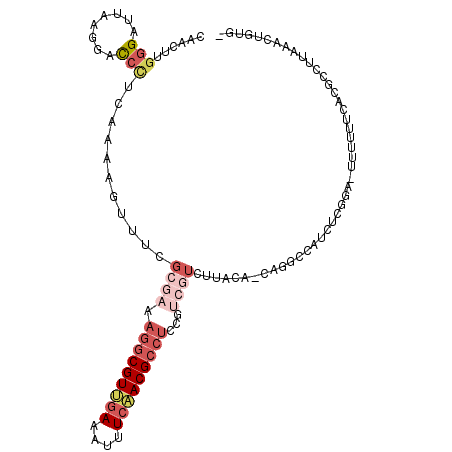
>X_DroMel_CAF1 2539253 108 + 22224390 CAACUUGGGAUUAAGGUUCUUCAAAAGUUUCGCGAAAGGCGUUGAAAUUUCAACGCCUUCGUCGUCCUACAACGGGCCAUCUCCGA-UUUUUUCACGCCUUAAACUGUG- .......((.(((((((.....(((((((..(((((.((((((((....))))))))))))).((((......)))).......))-)))))....))))))).))...- ( -31.80) >DroPse_CAF1 2359 89 + 1 -------AGAAGAAGGUCCGGCAAAACUUUU-CCAAAAGCGUGAAAAUUUCCACGGA------------AA-UUUGCCUACGCGGAUUCCCUUCACGCCCCAUGCCACGC -------.((((..(((((((.((....)).-))....(((((.(((((((....))------------))-)))...))))))))))..))))................ ( -23.00) >DroSec_CAF1 2620 107 + 1 CAACUUGGGAUUAUGGACCCUCAAAAGUUUCGCGAAAGGCGUUGAAAUUUCAACGCCUCCGUCGUCUUACA-CAGGCCAUCUCGGA-UUUUUUCACGCCUUAAACUGUG- .((((((((........)))....)))))..((((.(((((((((....)))))))))...))))....((-(((.......((((-.....)).)).......)))))- ( -28.74) >DroSim_CAF1 2609 107 + 1 CAACUUGGGUUUAAGGACCCUCAAAAGUUUCGCGAAAGGCGUUGAAAUUUCAACGCCUCCGUCGUCUUACA-CAGGCCAUCUCGGA-UUUUUUCACGCCCCAAACUGUG- .((((((((((....)))))....)))))..((((.(((((((((....)))))))))...))))....((-(((.......((((-.....)).)).......)))))- ( -31.74) >DroEre_CAF1 2791 105 + 1 CAACUUGGGAUUAAG-GUCCGUCAAAGUUUCGCGAAAGGCGUUGAAAUUUCCACGCCUCCGUCGUCUUGCA-CUGGACUGCUCG-C-CUUUUUCACGCCUUGAACUGUG- .......((.(((((-(..(((.((((....((((.((((((.((....)).))))))..((.((((....-..)))).)))))-)-..)))).))))))))).))...- ( -28.00) >DroYak_CAF1 2835 88 + 1 CAACUUGGGAUUAAGGGUCCUGCAAAGUUUCUCGAAAGGCGUUGAAAUUUCAACGCCUCCUC------------GGAC---------UUUUUUCACGCCUUAAACAGUG- ..(((.(...((((((((.....((((((...(((.(((((((((....)))))))))..))------------))))---------)))....)).)))))).)))).- ( -25.50) >consensus CAACUUGGGAUUAAGGACCCUCAAAAGUUUCGCGAAAGGCGUUGAAAUUUCAACGCCUCCGUCGUCUUACA_CAGGCCAUCUCGGA_UUUUUUCACGCCUUAAACUGUG_ ......(((........)))...........((((.(((((((((....)))))))))...))))............................................. (-11.11 = -12.90 + 1.79)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:01 2006