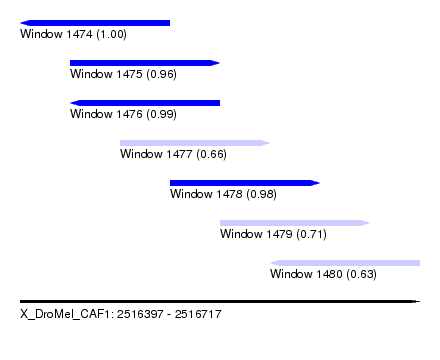
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,516,397 – 2,516,717 |
| Length | 320 |
| Max. P | 0.998236 |

| Location | 2,516,397 – 2,516,517 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -54.08 |
| Consensus MFE | -47.08 |
| Energy contribution | -48.00 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2516397 120 - 22224390 AAUAUAUGUGACCGUCGGUGGCCAGGCUCUUGGCAUCGCGUCGACGUGCCAUGGCCGCCGCCACUGUGAGUUCCGCCUGAUCCGGUGGCAAAAGUCGGUUCUUCGGCUUGGCAUCCACCU .......(((...((((((((((((((.((((((.....))))).).))).))))))))(((((((.((...........)))))))))..(((((((....))))))))))...))).. ( -51.10) >DroSec_CAF1 14599 120 - 1 AAUAUAUGUGGCCGUCGGUGGCCAGGCUCUUGGCAUCGCGUCGACGUGCCAUGGCCGCCGCCACUGUGAGUUCCGCCUGAUCCGGUGGCAGAAGUCGGUUCUUCGGCUUGGCAUCCACCU .......((((..((((((((((((((.((((((.....))))).).))).))))))))(((((((.((...........)))))))))..(((((((....))))))))))..)))).. ( -55.00) >DroSim_CAF1 14588 120 - 1 AAUAUAUGUGGCCGUCGGUGGCCAGGCUCUUGGCAUCGCGUCGACGUGCCAUGGCCGCCGCCACUGUGAGUUCCGCCUGAUCCGGUGGCAGAAGCCGGUUCUUCGGCUUGGCAUCCACCU .......((((..((((((((((((((.((((((.....))))).).))).))))))))(((((((.((...........)))))))))..(((((((....))))))))))..)))).. ( -57.70) >DroEre_CAF1 15384 120 - 1 AAUAUAUGUGACCGUCGGUGGCCAGGCUCUUGGCAUCCCGUCGACGGGCAAUGGCCGCCGCCACUGUCAGUUCCGCCUGAUCCGGUGGCAGAAGUCGAUUUUUCGGCUUGGCAUCCAUCA .....(((.((..(((((((((((.(((((((((.....))))).))))..))))))))(((((((((((......))))..)))))))..(((((((....)))))))))).))))).. ( -53.10) >DroYak_CAF1 16623 120 - 1 AAUAGAUGUGGCCAUCGGUGGCCAGGCUCUUGGCAUCCCGGCGACGGGCCAUUGCCGCCGCCACUGUGAGUUCCGCCUGAUCCGGUGGCAGAAGUCGGUUUUUCGGCUUGGCAUCCACCU ....((((....))))(((((...(((...((((...(((....)))))))..)))((((((((((.((...........)))))))))..(((((((....))))))))))..))))). ( -53.50) >consensus AAUAUAUGUGGCCGUCGGUGGCCAGGCUCUUGGCAUCGCGUCGACGUGCCAUGGCCGCCGCCACUGUGAGUUCCGCCUGAUCCGGUGGCAGAAGUCGGUUCUUCGGCUUGGCAUCCACCU .......((((..((((((((((((((.((((((.....))))).).))).))))))))(((((((.((...........)))))))))..(((((((....))))))))))..)))).. (-47.08 = -48.00 + 0.92)

| Location | 2,516,437 – 2,516,557 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -47.50 |
| Consensus MFE | -41.68 |
| Energy contribution | -42.04 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2516437 120 + 22224390 UCAGGCGGAACUCACAGUGGCGGCGGCCAUGGCACGUCGACGCGAUGCCAAGAGCCUGGCCACCGACGGUCACAUAUAUUUCCCACUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUC ....(((((.(((..((((((((.(((((((((((((....))).)))))......))))).)))..(....).........)))))(((.......))))))..))))).......... ( -42.60) >DroSec_CAF1 14639 120 + 1 UCAGGCGGAACUCACAGUGGCGGCGGCCAUGGCACGUCGACGCGAUGCCAAGAGCCUGGCCACCGACGGCCACAUAUAUUUCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUC ....(((((.......(((((((.(((((((((((((....))).)))))......))))).))....))))).......((((((((........))))).)))))))).......... ( -48.00) >DroSim_CAF1 14628 120 + 1 UCAGGCGGAACUCACAGUGGCGGCGGCCAUGGCACGUCGACGCGAUGCCAAGAGCCUGGCCACCGACGGCCACAUAUAUUUCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUC ....(((((.......(((((((.(((((((((((((....))).)))))......))))).))....))))).......((((((((........))))).)))))))).......... ( -48.00) >DroEre_CAF1 15424 120 + 1 UCAGGCGGAACUGACAGUGGCGGCGGCCAUUGCCCGUCGACGGGAUGCCAAGAGCCUGGCCACCGACGGUCACAUAUAUUUCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUC ....(((((..((((..(((.((((((..(((((((....))))....)))..)))..))).)))...))))........((((((((........))))).)))))))).......... ( -45.30) >DroYak_CAF1 16663 120 + 1 UCAGGCGGAACUCACAGUGGCGGCGGCAAUGGCCCGUCGCCGGGAUGCCAAGAGCCUGGCCACCGAUGGCCACAUCUAUUUCCCGCUGCUGAAGAUCAGCGAGGAUCCGCACAUUGAUGC ....(((((.(((..((((((((((((....).))))))))((((.....(((...((((((....))))))..)))...)))))))((((.....)))))))..))))).......... ( -53.60) >consensus UCAGGCGGAACUCACAGUGGCGGCGGCCAUGGCACGUCGACGCGAUGCCAAGAGCCUGGCCACCGACGGCCACAUAUAUUUCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUC ....(((((.......(((((((.(((((.(((.((((.....))))......)))))))).))....))))).......((((((((........))))).)))))))).......... (-41.68 = -42.04 + 0.36)

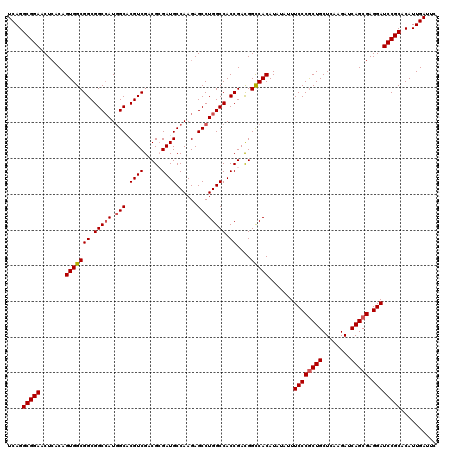
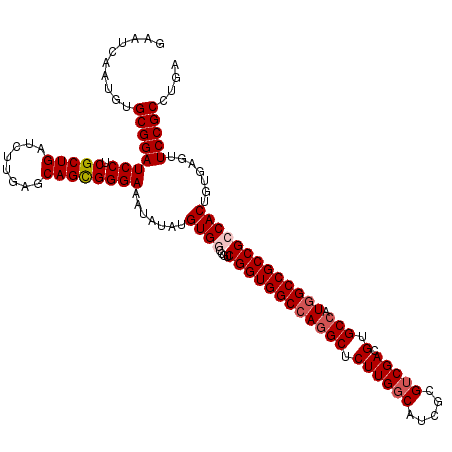
| Location | 2,516,437 – 2,516,557 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -52.42 |
| Consensus MFE | -46.62 |
| Energy contribution | -47.46 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2516437 120 - 22224390 GAAUCAAUGUGCGGAUCCUCGCUGAUCUUGAGCAGUGGGAAAUAUAUGUGACCGUCGGUGGCCAGGCUCUUGGCAUCGCGUCGACGUGCCAUGGCCGCCGCCACUGUGAGUUCCGCCUGA ..........((((((((.(((((........))))))))..((((.(((...(.((((((((((((.((((((.....))))).).))).)))))))))))))))))...))))).... ( -48.50) >DroSec_CAF1 14639 120 - 1 GAAUCAAUGUGCGGAUCCUCGCUGAUCUUGAGCAGCGGGAAAUAUAUGUGGCCGUCGGUGGCCAGGCUCUUGGCAUCGCGUCGACGUGCCAUGGCCGCCGCCACUGUGAGUUCCGCCUGA ..........((((((((.(((((........))))))))..((((.(((((....(((((((((((.((((((.....))))).).))).)))))))))))))))))...))))).... ( -54.70) >DroSim_CAF1 14628 120 - 1 GAAUCAAUGUGCGGAUCCUCGCUGAUCUUGAGCAGCGGGAAAUAUAUGUGGCCGUCGGUGGCCAGGCUCUUGGCAUCGCGUCGACGUGCCAUGGCCGCCGCCACUGUGAGUUCCGCCUGA ..........((((((((.(((((........))))))))..((((.(((((....(((((((((((.((((((.....))))).).))).)))))))))))))))))...))))).... ( -54.70) >DroEre_CAF1 15424 120 - 1 GAAUCAAUGUGCGGAUCCUCGCUGAUCUUGAGCAGCGGGAAAUAUAUGUGACCGUCGGUGGCCAGGCUCUUGGCAUCCCGUCGACGGGCAAUGGCCGCCGCCACUGUCAGUUCCGCCUGA ..........((((((((.(((((........))))))))........((((.(((((((((((.(((((((((.....))))).))))..)))))))))..)).))))..))))).... ( -52.30) >DroYak_CAF1 16663 120 - 1 GCAUCAAUGUGCGGAUCCUCGCUGAUCUUCAGCAGCGGGAAAUAGAUGUGGCCAUCGGUGGCCAGGCUCUUGGCAUCCCGGCGACGGGCCAUUGCCGCCGCCACUGUGAGUUCCGCCUGA ..........(((((..(((((((.....)))).((((......((((....))))((((((..(((...((((...(((....)))))))..))))))))).))))))).))))).... ( -51.90) >consensus GAAUCAAUGUGCGGAUCCUCGCUGAUCUUGAGCAGCGGGAAAUAUAUGUGGCCGUCGGUGGCCAGGCUCUUGGCAUCGCGUCGACGUGCCAUGGCCGCCGCCACUGUGAGUUCCGCCUGA ..........((((((((.(((((........)))))))).......((((....((((((((((((.((((((.....))))).).))).))))))))))))).......))))).... (-46.62 = -47.46 + 0.84)

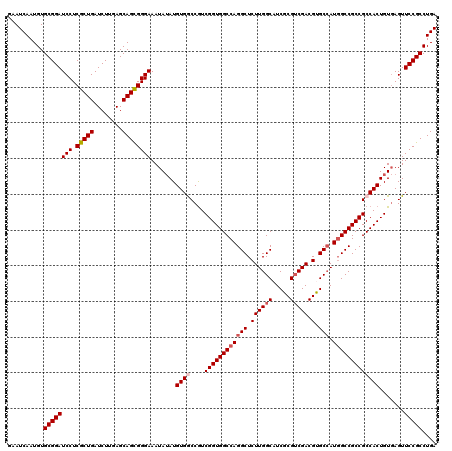
| Location | 2,516,477 – 2,516,597 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -36.42 |
| Energy contribution | -36.38 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2516477 120 + 22224390 CGCGAUGCCAAGAGCCUGGCCACCGACGGUCACAUAUAUUUCCCACUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUG .(((((.....((((.(((((......)))))...............))))..(((((((..(((((........)))))...))))))).)))))..((.((.((....)).))))... ( -37.29) >DroSec_CAF1 14679 120 + 1 CGCGAUGCCAAGAGCCUGGCCACCGACGGCCACAUAUAUUUCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUG .(((((.....((((.(((((......)))))...............))))..(((((((..(((((........)))))...))))))).)))))..((.((.((....)).))))... ( -39.99) >DroSim_CAF1 14668 120 + 1 CGCGAUGCCAAGAGCCUGGCCACCGACGGCCACAUAUAUUUCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUG .(((((.....((((.(((((......)))))...............))))..(((((((..(((((........)))))...))))))).)))))..((.((.((....)).))))... ( -39.99) >DroEre_CAF1 15464 120 + 1 CGGGAUGCCAAGAGCCUGGCCACCGACGGUCACAUAUAUUUCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUG ..((.((((..((((((((((......)))))..........((..(((....(((((((..(((((........)))))...)))))))....))).))..)).)))..)).))))... ( -38.20) >DroYak_CAF1 16703 120 + 1 CGGGAUGCCAAGAGCCUGGCCACCGAUGGCCACAUCUAUUUCCCGCUGCUGAAGAUCAGCGAGGAUCCGCACAUUGAUGCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUG ..(((.(((((((...((((((....))))))..))).....((..(((.((.(((((((..(.(((........))).)...)))))))..))))).)).)))).)))........... ( -41.90) >consensus CGCGAUGCCAAGAGCCUGGCCACCGACGGCCACAUAUAUUUCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUG ...(.((((..((((((((((......)))))..........((..(((....(((((((..(((((........)))))...)))))))....))).))..)).)))..)).)).)... (-36.42 = -36.38 + -0.04)

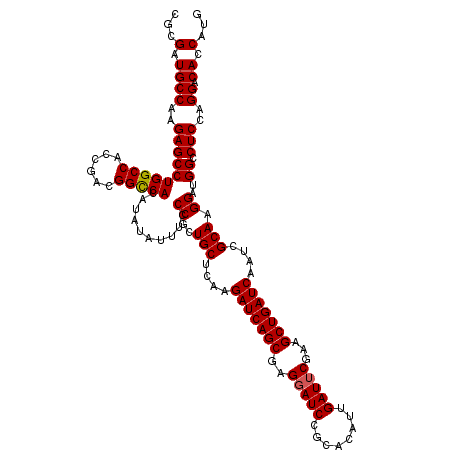
| Location | 2,516,517 – 2,516,637 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -41.82 |
| Consensus MFE | -38.64 |
| Energy contribution | -39.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2516517 120 + 22224390 UCCCACUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAAUUCGGCAGUCCAAAGUUGCGAGAGA ........(((..(((((((..(((((........)))))...)))))))..((((((((.((.((....)).))))........(((((((..........))))))).))))))))). ( -40.40) >DroSec_CAF1 14719 120 + 1 UCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAGUUGCGGGAGA ((((((.(((...(((((((..(((((........)))))...)))))))........((((.(((....((((.(((.........)))....))))))).))))..))).)))))).. ( -42.60) >DroSim_CAF1 14708 120 + 1 UCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAGUUGCGGGAGA ((((((.(((...(((((((..(((((........)))))...)))))))........((((.(((....((((.(((.........)))....))))))).))))..))).)))))).. ( -42.60) >DroEre_CAF1 15504 120 + 1 UCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAAUUGCGGGAGA (((((((((....(((((((..(((((........)))))...)))))))....))).((((.(((....((((.(((.........)))....))))))).))))......)))))).. ( -42.10) >DroYak_CAF1 16743 120 + 1 UCCCGCUGCUGAAGAUCAGCGAGGAUCCGCACAUUGAUGCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAGUUGCGGGAGA ((((((.(((...(((((((..(.(((........))).)...)))))))........((((.(((....((((.(((.........)))....))))))).))))..))).)))))).. ( -41.40) >consensus UCCCGCUGCUCAAGAUCAGCGAGGAUCCGCACAUUGAUUCGAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAGUUGCGGGAGA (((((((((....(((((((..(((((........)))))...)))))))....))).((((.(((((((((((.....))....)))))).(....)))).))))......)))))).. (-38.64 = -39.24 + 0.60)

| Location | 2,516,557 – 2,516,677 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.58 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -37.92 |
| Energy contribution | -38.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2516557 120 + 22224390 GAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAAUUCGGCAGUCCAAAGUUGCGAGAGAAGUUCGCCCGCAAGCAGAAGCAGCUGCUCGCCAAGCAGCA ...((((.((..((((((((.((.((....)).))))........(((((((..........))))))).))))))..)).........((.(((((......))))).))....)))). ( -37.30) >DroSec_CAF1 14759 120 + 1 GAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAGUUGCGGGAGAAGUUCGCCCGCAAGCAGAAGCAGCUGCUCGCCAAGCAGCA ...((((.......(((.((.((.((....)).)))).)))....(((((.(((((..(((.........(((((((.(.....).)))))))((....)).))))))))))))))))). ( -41.90) >DroSim_CAF1 14748 120 + 1 GAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAGUUGCGGGAGAAGUUCGCCCGCAAGCAGAAGCAGCUGCUCGCCAAGCAGCA ...((((.......(((.((.((.((....)).)))).)))....(((((.(((((..(((.........(((((((.(.....).)))))))((....)).))))))))))))))))). ( -41.90) >DroEre_CAF1 15544 120 + 1 GAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAAUUGCGGGAGAAGUUCGCCCGCAAGCAGAAGCAACUGCUAGCCAAGCAGCA ...((((.....((....)).((((.....((((.(((.........))).((....))...)))).....((((((.(.....).))))))(((((......))))).))))..)))). ( -39.40) >DroYak_CAF1 16783 120 + 1 GAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAGUUGCGGGAGAAGUUCGCCCGCAAGCAGAAGCAACUGCUUGCCAAGCAGCA ...(((((....(((...(((.....)))......(((.........))).)))..)))))........((((((((.(.....).)))((((((((......))))))))...))))). ( -39.60) >consensus GAAGCUGAUCAAUCGCAAGGAUGGCCUCCAGGACACCAUGUAUUAUUUGGACGAGUUCGGCAGUCCCAAGUUGCGGGAGAAGUUCGCCCGCAAGCAGAAGCAGCUGCUCGCCAAGCAGCA ...((((.....((....)).((((.....((((.(((.........))).((....))...))))....(((((((.(.....).)))))))((((......))))..))))..)))). (-37.92 = -38.12 + 0.20)

| Location | 2,516,597 – 2,516,717 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -27.36 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2516597 120 - 22224390 CUGCUCCUCGCUCCUCCUUUCACGUUUCAUCAACUGCUUCUGCUGCUUGGCGAGCAGCUGCUUCUGCUUGCGGGCGAACUUCUCUCGCAACUUUGGACUGCCGAAUUCGUCCAAAUAAUA .......((((((.................((((.((....)).).)))((((((((......))))))))))))))..............(((((((..........)))))))..... ( -30.40) >DroSec_CAF1 14799 120 - 1 CUGCUCCUCGCUCCUCCUCUCCCGUUUCAUCAUCUGCUUCUGCUGCUUGGCGAGCAGCUGCUUCUGCUUGCGGGCGAACUUCUCCCGCAACUUGGGACUGCCGAACUCGUCCAAAUAAUA ..(((.((((((...........(......)....((....)).....)))))).))).((....))(((((((.(.....).)))))))....((((..........))))........ ( -29.80) >DroSim_CAF1 14788 120 - 1 CUGCUCCUCGCUCCUCCGCUCCCGUUUCAUCAUCUGCUUCUGCUGCUUGGCGAGCAGCUGCUUCUGCUUGCGGGCGAACUUCUCCCGCAACUUGGGACUGCCGAACUCGUCCAAAUAAUA ..((.....))...((.(((((((...........((....(((((((...))))))).))......(((((((.(.....).)))))))..)))))..)).))................ ( -31.90) >DroEre_CAF1 15584 120 - 1 CUGCUCCUCGCUCCUUCUCUCCCGUUUCAUCAACUGCUUCUGCUGCUUGGCUAGCAGUUGCUUCUGCUUGCGGGCGAACUUCUCCCGCAAUUUGGGACUGCCGAACUCGUCCAAAUAAUA ..((.....))........((((((.....((((((((...(((....))).)))))))).....))(((((((.(.....).)))))))...))))....................... ( -33.70) >DroYak_CAF1 16823 120 - 1 CUGCUCCUCGCUCCUUCGCUCCCGCUUCAUCAUCUGCUUCUGCUGCUUGGCAAGCAGUUGCUUCUGCUUGCGGGCGAACUUCUCCCGCAACUUGGGACUGCCGAACUCGUCCAAAUAAUA ..((.....))...(((((((((((.....((.(((((..((((....))))))))).)).....))(((((((.(.....).)))))))...))))..).))))............... ( -33.40) >consensus CUGCUCCUCGCUCCUCCUCUCCCGUUUCAUCAUCUGCUUCUGCUGCUUGGCGAGCAGCUGCUUCUGCUUGCGGGCGAACUUCUCCCGCAACUUGGGACUGCCGAACUCGUCCAAAUAAUA ..((.....))............((.....((.((((((..(((....))))))))).)).....))(((((((.(.....).)))))))....((((..........))))........ (-27.36 = -27.12 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:55 2006