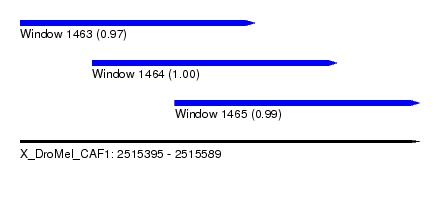
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,515,395 – 2,515,589 |
| Length | 194 |
| Max. P | 0.999272 |

| Location | 2,515,395 – 2,515,509 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.97 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.16 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2515395 114 + 22224390 AUAUACCCA----A-AACAGAAAAACUGUGCCGCUUUAGACGCUUUAUCAAUUUCAAAGAACCGAAAAGGAAAUACUCUA-ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGG .........----.-..........((((.((((.......))........((((........)))).))((((((((((-.....)))))))))).)))).((((((......)))))) ( -21.10) >DroSec_CAF1 13619 113 + 1 AU--ACCCA----AAAACAGAAAAACUGUGCCGCUUUAGACGCUUUGUAAGUUUCAAAGAACCGAAAAGGAAAUACUCUA-ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGG ..--.....----............((((.((..((..(...(((((.......)))))..)..))..))((((((((((-.....)))))))))).)))).((((((......)))))) ( -24.00) >DroSim_CAF1 13609 112 + 1 AU--ACCCA----A-AACAGAAAAACUGUGCCGCUUUAGACGCUUUAUACGUUUCAAAGAACCGAAAAGGAAAUACUCUA-ACGCCUAGAGUAUUCAACAGACCAUUAAAAACCUGAUGG ..--.....----.-..........((((....(((((((((.......))))).))))..((.....)).(((((((((-.....)))))))))..)))).((((((......)))))) ( -22.00) >DroEre_CAF1 14342 118 + 1 GU--AUCCAUACAAAUCCUGAAAAACUGUGCCGCUUAAGACGCUUGAUAAGUUUUUUAAAACCGAAAAGGAAAUACUCUAAACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGG ..--..............(((((((((...(.((.......))..)...)))))))))...((.....))((((((((((......))))))))))......((((((......)))))) ( -22.70) >DroYak_CAF1 15595 113 + 1 GU--ACC-AU---CAUACUGAAAAACUGUGCCGCUUAAGACGCUUGAUAAGUUUUAAAGAACCGAAAGGGAAAUACUCUA-ACGCCUAGAGUAUUUAACAGACCAUUGAAAACCUGAUGG ..--.((-((---((..((..((((((...(.((.......))..)...))))))..))..((....)).((((((((((-.....))))))))))..................)))))) ( -26.40) >consensus AU__ACCCA____AAAACAGAAAAACUGUGCCGCUUUAGACGCUUUAUAAGUUUCAAAGAACCGAAAAGGAAAUACUCUA_ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGG .........................((((.((((.......))........((((........)))).))((((((((((......)))))))))).)))).((((((......)))))) (-19.36 = -19.16 + -0.20)

| Location | 2,515,430 – 2,515,549 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2515430 119 + 22224390 CGCUUUAUCAAUUUCAAAGAACCGAAAAGGAAAUACUCUA-ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACUACGCAGGCAGCAGGAGCUGCACCAGC .(((.......((((........)))).((((((((((((-.....))))))))))....(.((((((......)))))))...................(((((....))))).))))) ( -27.40) >DroSec_CAF1 13653 119 + 1 CGCUUUGUAAGUUUCAAAGAACCGAAAAGGAAAUACUCUA-ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACCACGCAGGCAGCAGGAGCUGCACCAGC .(((((((...((((........)))).((((((((((((-.....))))))))))....(.((((((......)))))))...........))..))))(((((....)))))...))) ( -29.30) >DroSim_CAF1 13642 119 + 1 CGCUUUAUACGUUUCAAAGAACCGAAAAGGAAAUACUCUA-ACGCCUAGAGUAUUCAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACCUCGCAGGGAGCAGGAGCUGCACCAGC .((........((((........))))(((.(((((((((-.....))))))))).....(.((((((......)))))))...........))).))..((.((((...)))).))... ( -25.30) >DroEre_CAF1 14380 120 + 1 CGCUUGAUAAGUUUUUUAAAACCGAAAAGGAAAUACUCUAAACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACCACGCAGGCAGCAGGAGCUGCACCAGC .((.((....(((........((.....))((((((((((......))))))))))))).(.((((((......)))))))............)).))..(((((....)))))...... ( -28.90) >DroYak_CAF1 15629 119 + 1 CGCUUGAUAAGUUUUAAAGAACCGAAAGGGAAAUACUCUA-ACGCCUAGAGUAUUUAACAGACCAUUGAAAACCUGAUGGCAACAACAACAACCACGCAGGCAGCAGGAGCUGCACCAGC .((.((....(((((......((....)).((((((((((-.....))))))))))............))))).((.((....)).)).....)).))..(((((....)))))...... ( -31.00) >consensus CGCUUUAUAAGUUUCAAAGAACCGAAAAGGAAAUACUCUA_ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACCACGCAGGCAGCAGGAGCUGCACCAGC .((........((((........)))).((((((((((((......))))))))))....(.((((((......)))))))...........))..))..(((((....)))))...... (-25.50 = -25.34 + -0.16)

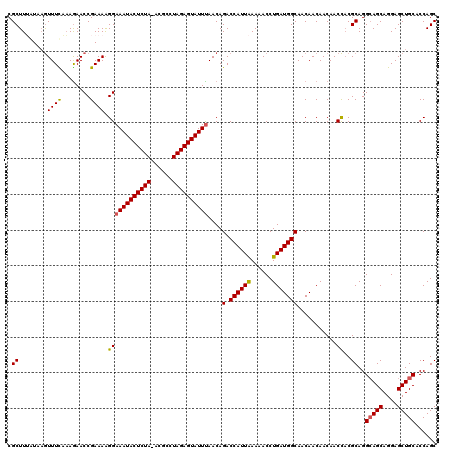
| Location | 2,515,470 – 2,515,589 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2515470 119 + 22224390 -ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACUACGCAGGCAGCAGGAGCUGCACCAGCUCUCAAUUUAUUGCCCGCCAGCAAUAACAAUAUAAAUAAU -..((((.((((..........((((((......))))))...........))).)..))))....(((((((...)))))))....(((((((......)))))))............. ( -25.60) >DroSec_CAF1 13693 119 + 1 -ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACCACGCAGGCAGCAGGAGCUGCACCAGCUCUCAAUUUAUUGCCCGCCAGCAAUAACCACAUAAAUAAU -..((((...((.....)).(.((((((......))))))).................))))....(((((((...)))))))....(((((((......)))))))............. ( -25.00) >DroSim_CAF1 13682 119 + 1 -ACGCCUAGAGUAUUCAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACCUCGCAGGGAGCAGGAGCUGCACCAGCUCUCAAUCUAUUGCCCGCCAGCAAUAACAACAUAAAUAAU -...(((.(((...((....))((((((......)))))).............)))..)))((...(((((((...)))))))...))((((((......)))))).............. ( -24.20) >DroEre_CAF1 14420 120 + 1 AACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACCACGCAGGCAGCAGGAGCUGCACCAGCUCUCAAUUUAUUGCCCGCCAGCAAUAAUAACACAAAUAAU ...((((...((.....)).(.((((((......))))))).................))))....(((((((...)))))))....(((((((......)))))))............. ( -25.10) >DroYak_CAF1 15669 116 + 1 -ACGCCUAGAGUAUUUAACAGACCAUUGAAAACCUGAUGGCAACAACAACAACCACGCAGGCAGCAGGAGCUGCACCAGCUCUCAACCUAUUGCCCGCCA---UUGAUAACACAAAUAAU -..((((((.((.(((((.......))))).)))))..)))...............((.(((((.((((((((...))))).....))).))))).))..---................. ( -23.50) >consensus _ACGCCUAGAGUAUUUAACAGACCAUUAAAAACCUGAUGGCAACAACAACAACCACGCAGGCAGCAGGAGCUGCACCAGCUCUCAAUUUAUUGCCCGCCAGCAAUAACAACAUAAAUAAU ...((((((.((.(((((.......))))).)))))..)))...............((.(((((..(((((((...))))))).......))))).))...................... (-20.60 = -20.88 + 0.28)

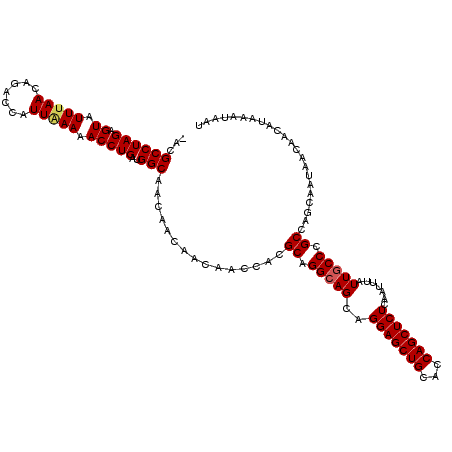
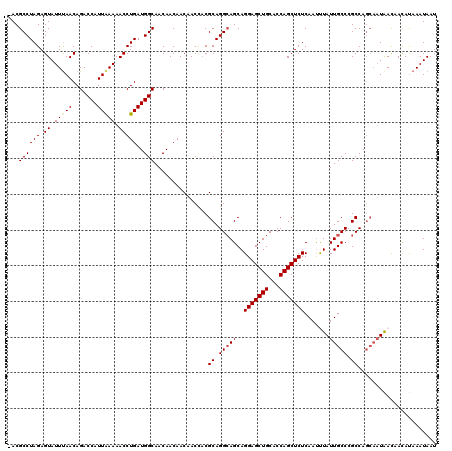
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:41 2006